The spectrIm-QMRS: Processing menu
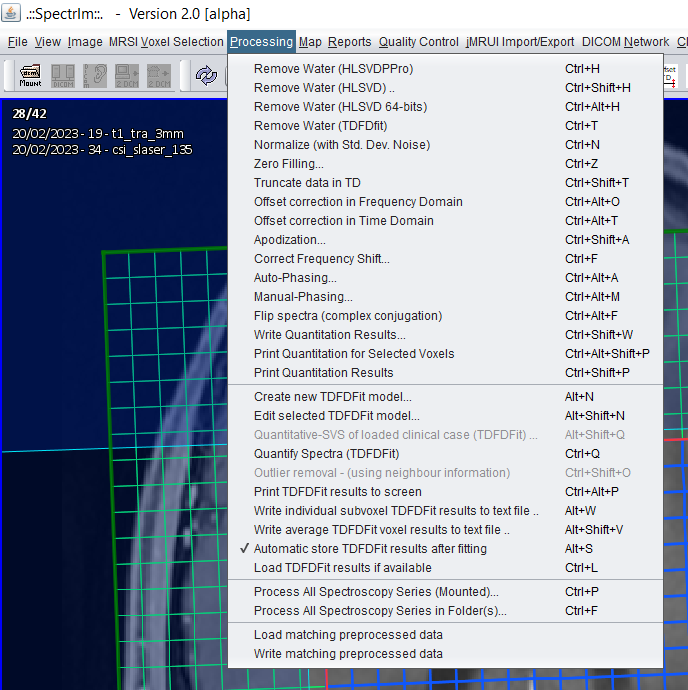
This menu enables the user to interactively perform processing and quantification of the loaded data.
The following view menu items are available:
- Create new TDFDFit model..
- Edit selected TDFDFit model..
- Quantitative-SVS of loaded clinical case (TDFDFit)..
- Quantify Spectra (TDFDFit)
- Outlier removal - (using neighbour information)..
- Print TDFDFit results to screen
- Write individual subvoxel TDFDFit results to text file ..
- Write average TDFDFit results to text file ..
- Automatically store TDFDFit results after fitting (toggle)
- Load TDFDFit results if available
- Process All Spectroscopy Series in Folder(s)
- Load matching preprocessed data
- Write matching preprocessed data
spectrIm-QMRS -> Processing -> Remove water (HSLVDpro)
Performs HLSVDpro directly on the selected spectra. In order to use this functionality, Python has to be installed. The default parameters used to perform the HLSVDpro water removal can defined after selecting:
"spectrIm-QMRS -> Processing -> Remove water (HSLVD).."
menu item, just underneath.
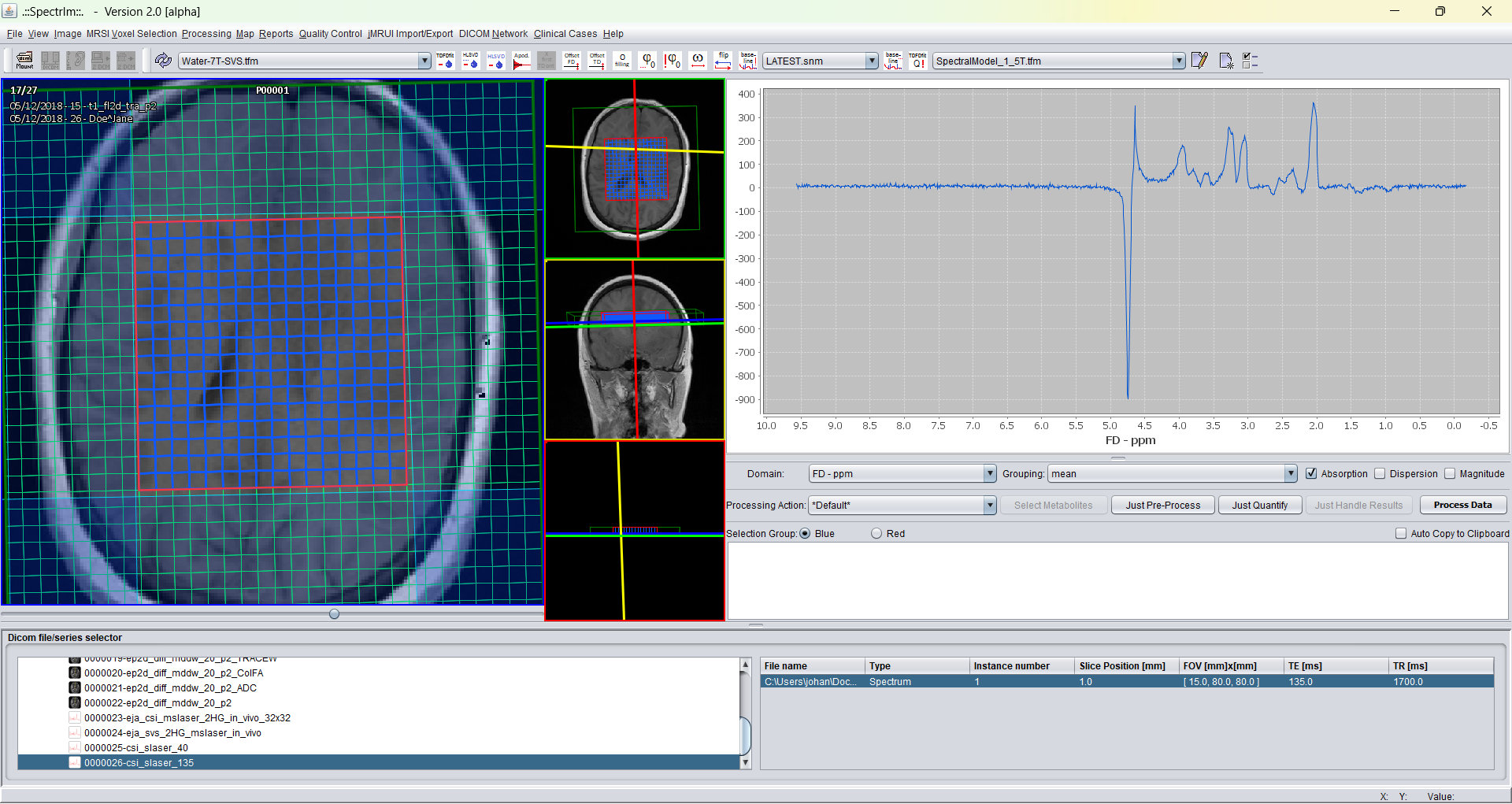
Example: before invoking HLSVDpro the loaded dataset looks following:
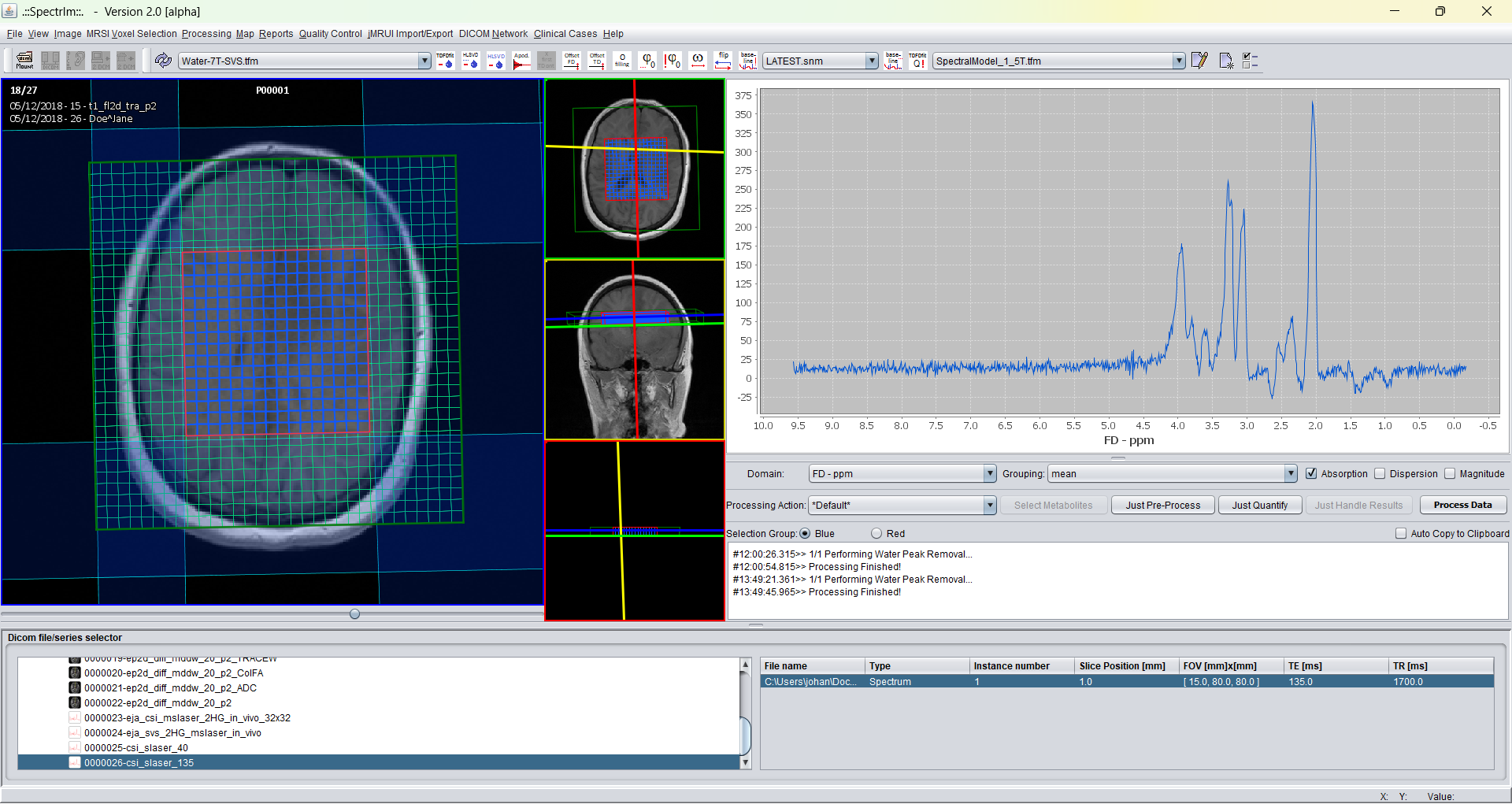
After loading and before performing water removal to be performed on the Siemens DICOM test dataset provided by the installation of spectrIm the GUI looks the following:
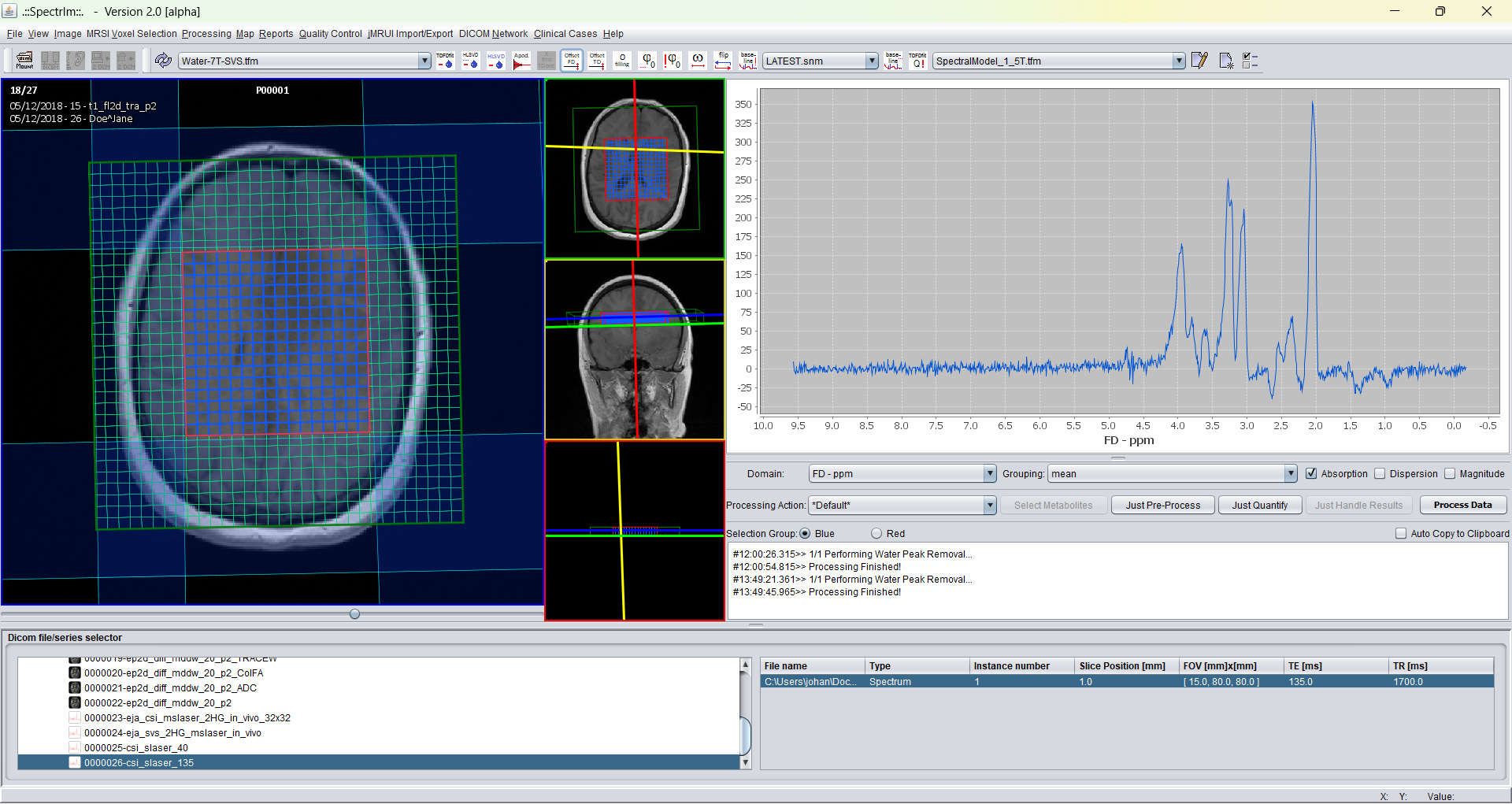
After performing HLSVDpro (which runs in parallel on the CPU of your computer; the more cores you have the faster this pre-processing job is done) the GUI looks the following:
Typically HLSVDpro of 256 spectra takes much less than a minute on a 8 core computer. The first inititialization of the Python server takes a large portion of computation time. Once the Python server is initialized, the computation is performed much faster.
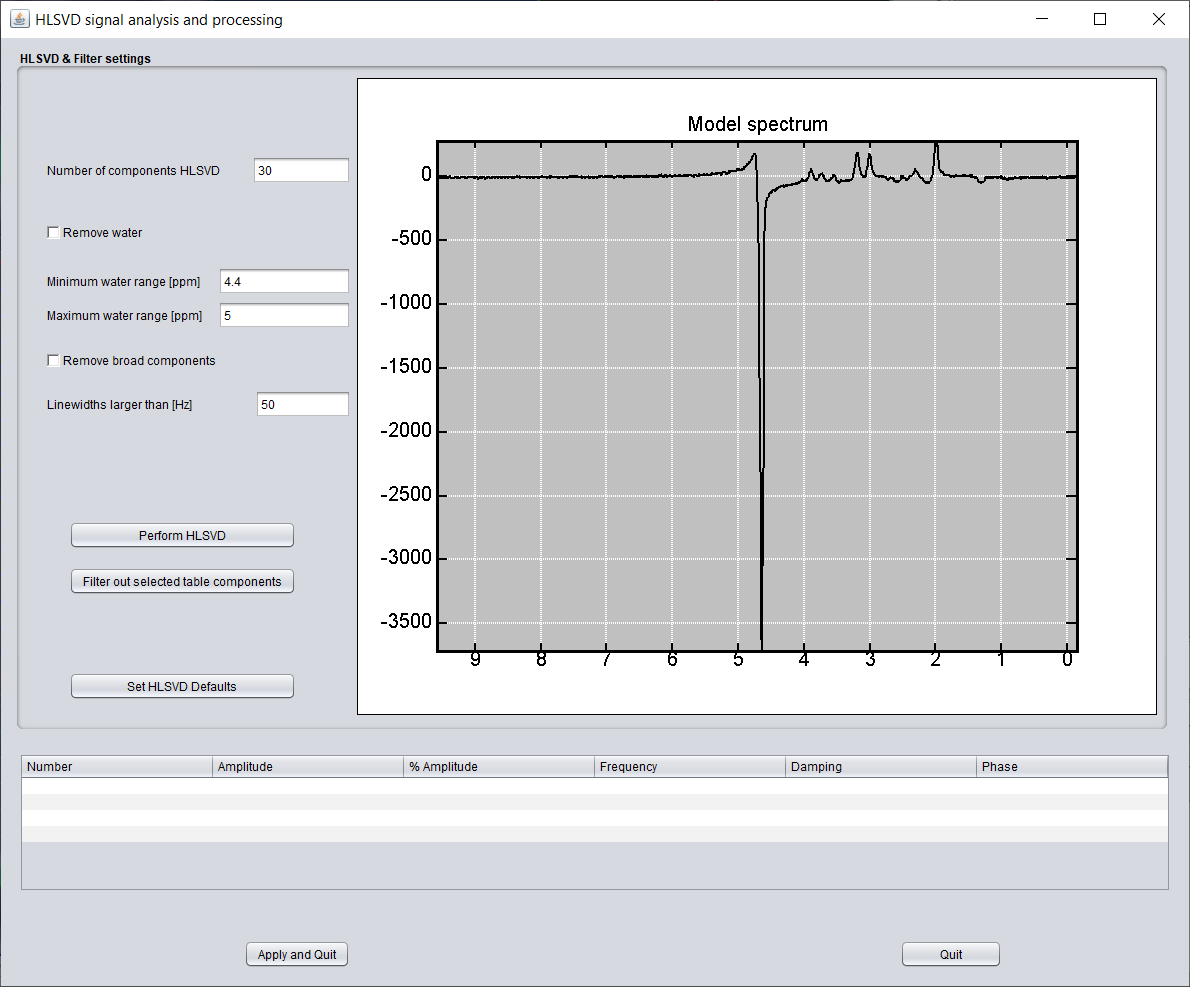
spectrIm-QMRS -> Processing -> Remove water (HSLVD)..
Opens a graphical user interface that enables the user to set default values on how HLSVD for water removal needs to be performed.

More information on this HLSVD settings GUI can be found Link to HLSVD-settings GUI
spectrIm-QMRS -> Processing -> Remove water (HSLVD 64 bits)..
Performs a 64-bits HLSVD algorithm on all user selected voxels. This is HLSVD routine is the 64 bits variant of the HLSVD 32-bit version of jMRUI-kernal. This routine uses the Java Native Interface ( JNI ) , and runs single threaded only. For 256 spectra is needs roughly 2 to 3 minutes, and is on an 8 cores computer factor 3-4 slower.
Similar to HLSVDpro, the performance of this algorithm is not always satisfactory for signals of which the oversampling of the raw scanner data is performed on the scanner by digital filtering. Sometimes the water removal may fail in this case. It is advice NOT to remove the oversampling of the spectral data to prevent running into this problem.
More information on this HLSVD settings GUI can be found Link to HLSVD-settings GUI
spectrIm-QMRS -> Processing -> Remove water (TDFDFit)
This function attempts to eliminate residual water (or any other spectral component(s)) by Prony's method. Prerequisite for using this function is the availability of a TDFDFit water-residual model. How to make such a model will be described elsewhere in this website ##H-link required here##.
After application of this TDFDFit based water removal assuming 1 water line the effect on the Siemens dataset is the following:
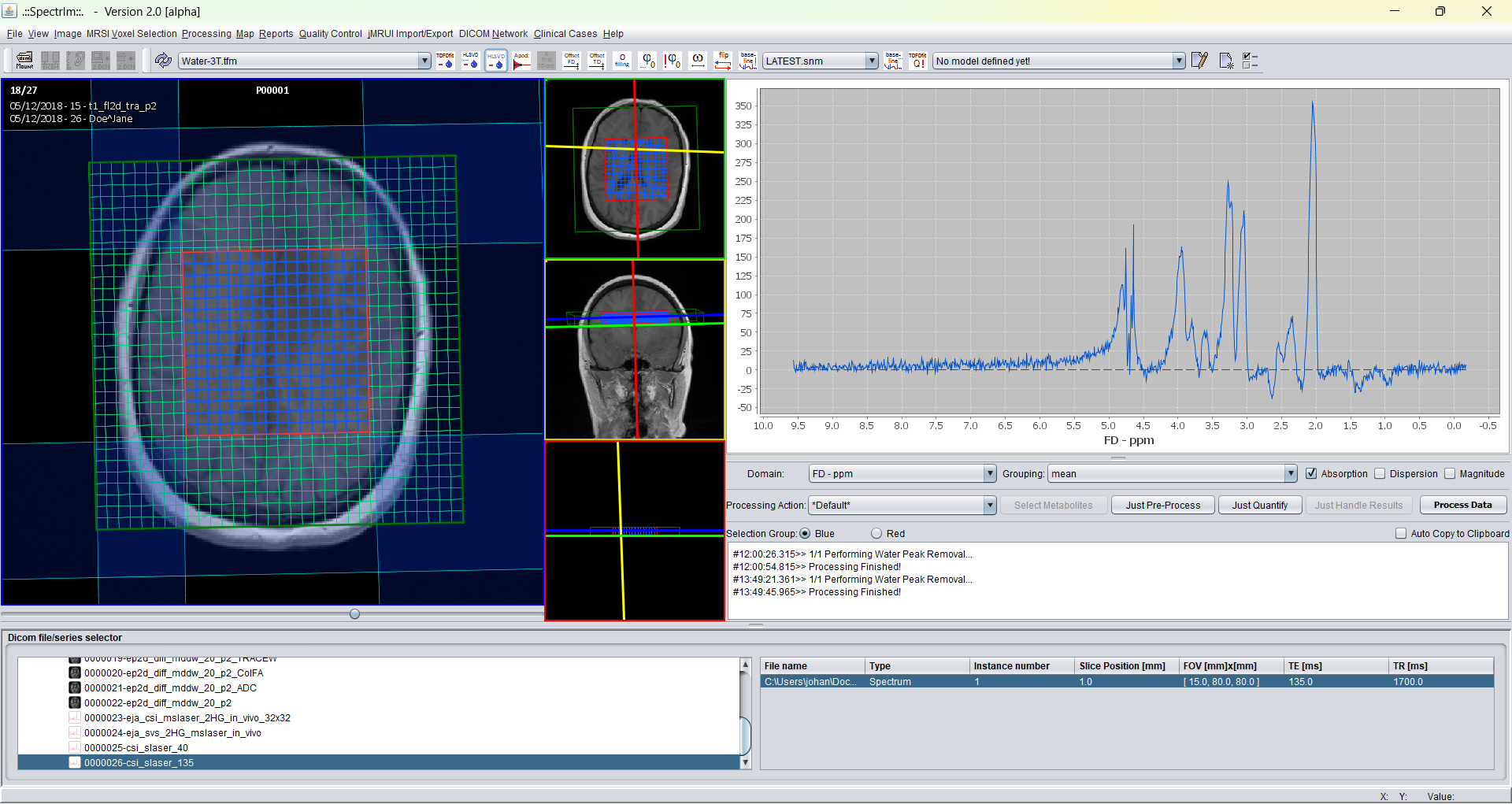
In the example displayed right above, only one single line was assumed to model the residual water, which is clearly too few.
This water removal is meant as fall-back option if HLSVDpro and HLSVD-64 bit fails to remove the residual water. Often, after applicatio of incomplete water removal using TDFDFit, HLSVDpro or HLSVD can successfully be applied.
spectrIm-QMRS -> Processing -> Normalize with St. Dev. Noise
This applies normalization to the complete loaded dataset. The standard deviation of the noise containing part of each individual response is computed, and each response is devided by this value.
This menu item is a implemented as checkbox. If selected the normalization is performed, if not checked the normalization is not performed. One can toggle between these states.
The normalization is applied independent on whether voxels of the dataset are selected or not.
As an example a spectrum of an 2D MRSI dataset without the noise normalization toggle selected:
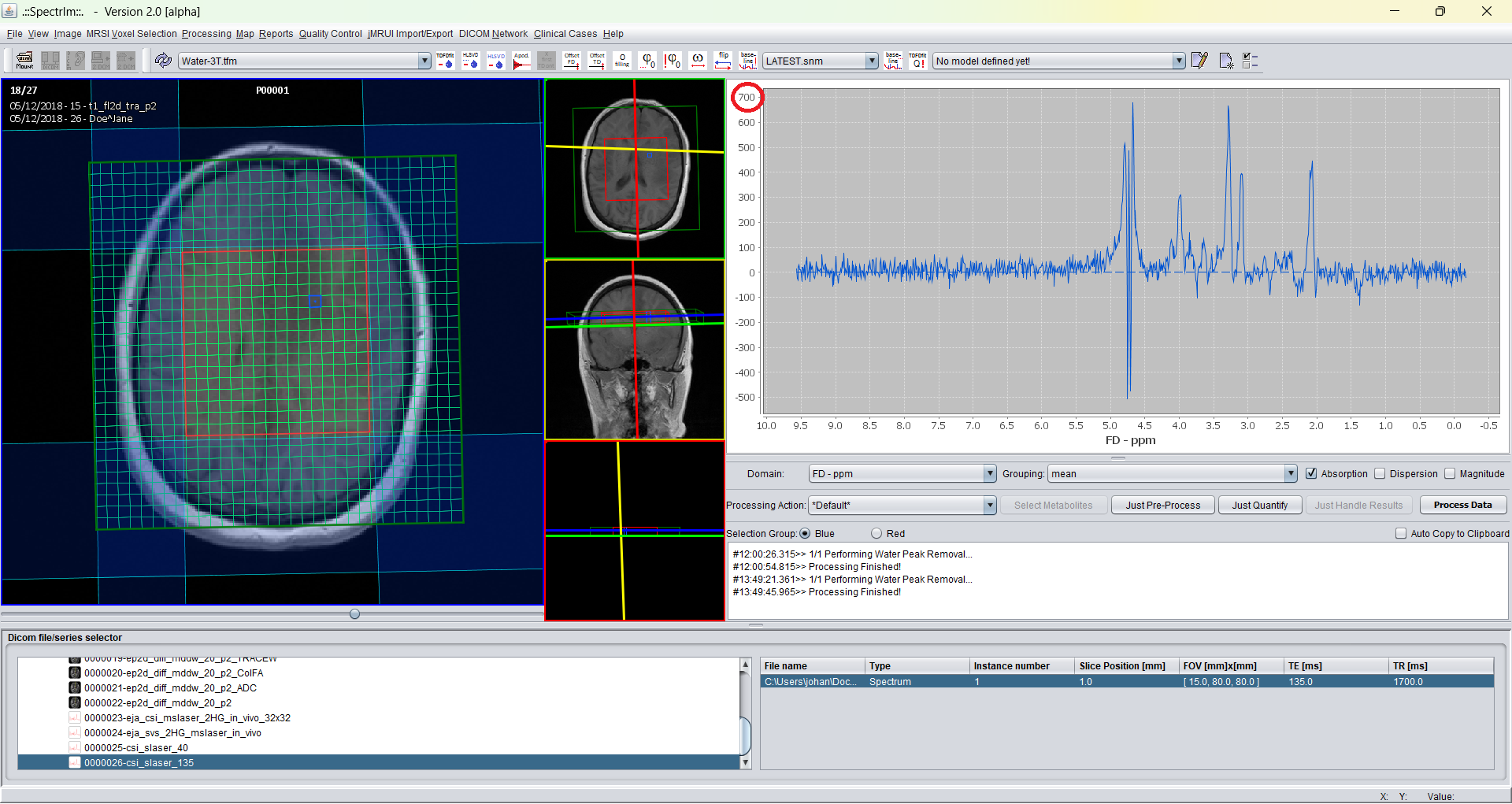
And with selecting noise level normalization. As expected the amplitude of the whole spectrum at a certain location is affected:
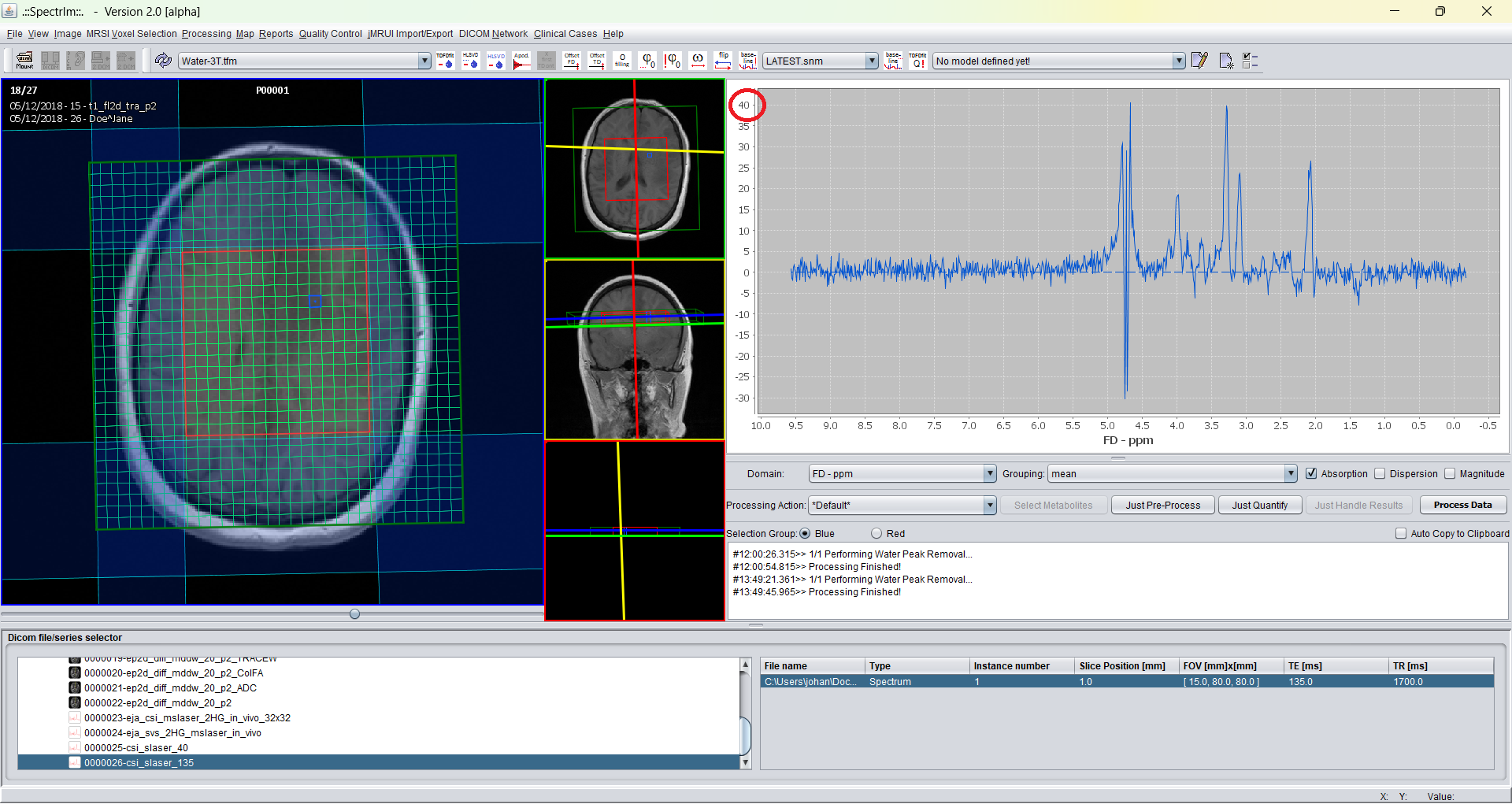
spectrIm-QMRS -> Processing -> Zero filling ..
Lets the user define the amount of points added in time domain in order to obtain an interpolation effect in frequency domain. This process is called zero filling. To get a deeper understanding of please refer to the literature (e.g. Andreas Ebel et al.). The number of points must be a power of 2, since the Fast Fourier Transformation used in spectrIm-QMRS requires this.
The popup dialog shown when envoking this menu item is following:
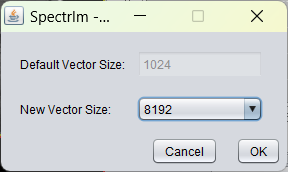
After applying zero filling from 1k (1024) to 8k points (8192) the spectrum desplayed above looks like following:
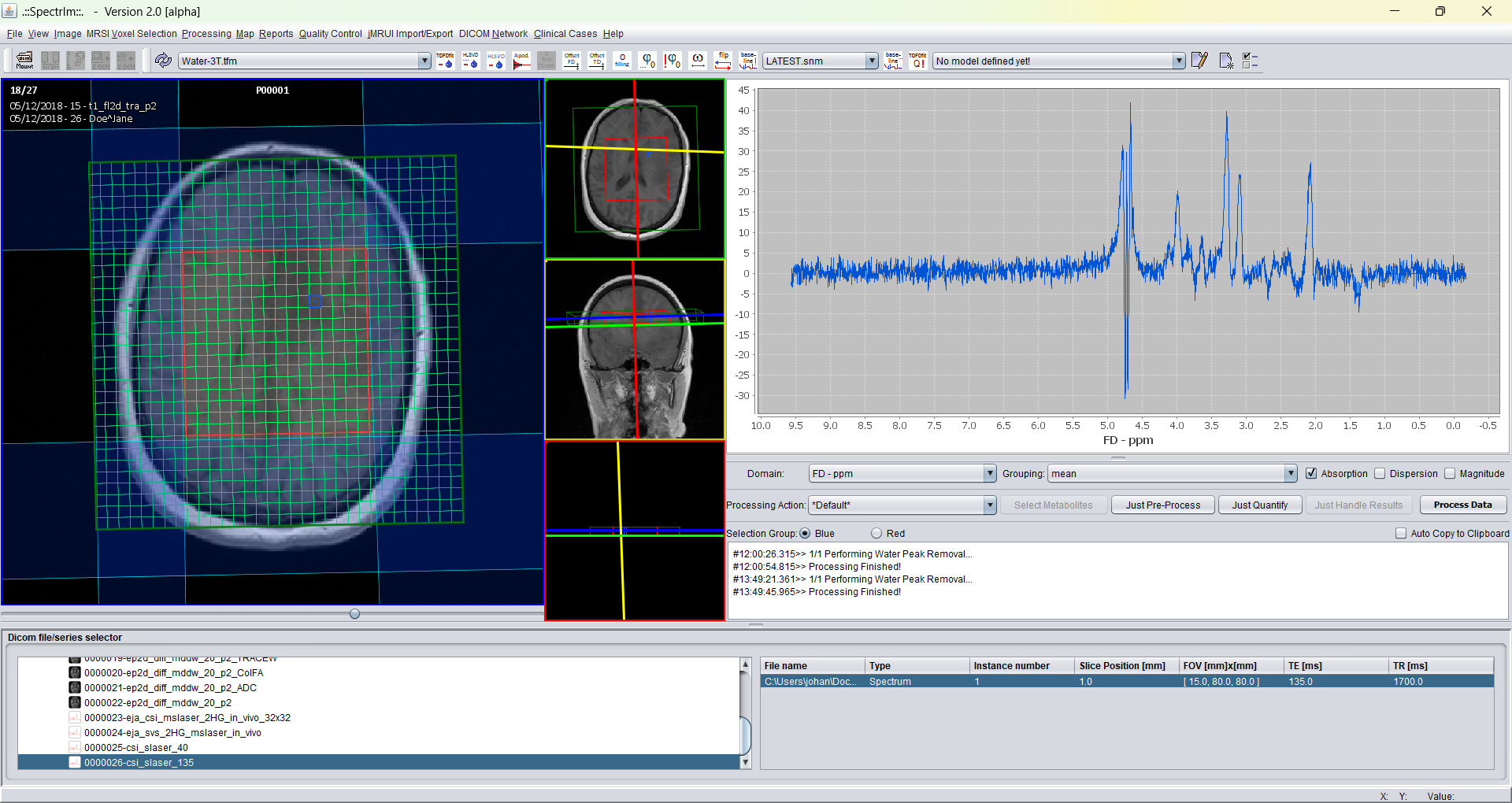
Please note that zero filling does not add information to the data. If zero filling is applied before quantification, the time required to quantify the data may be much longer, without adding anything to the accuracy of the data.
Additionally a pittfall should be mentioned namely that zero filling is performed without adeqaute apozation of the data, this may lead to artifacts to become visible in the spectra.
spectrIm-QMRS -> Processing -> Truncate data in time domain ..
Opens a graphical user interface that enables the user to set the new number of time domain points. The number of points must be a power of 2, since the Fast Fourier Transformation used in spectrIm-QMRS requires this.
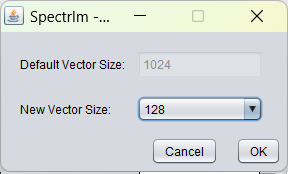
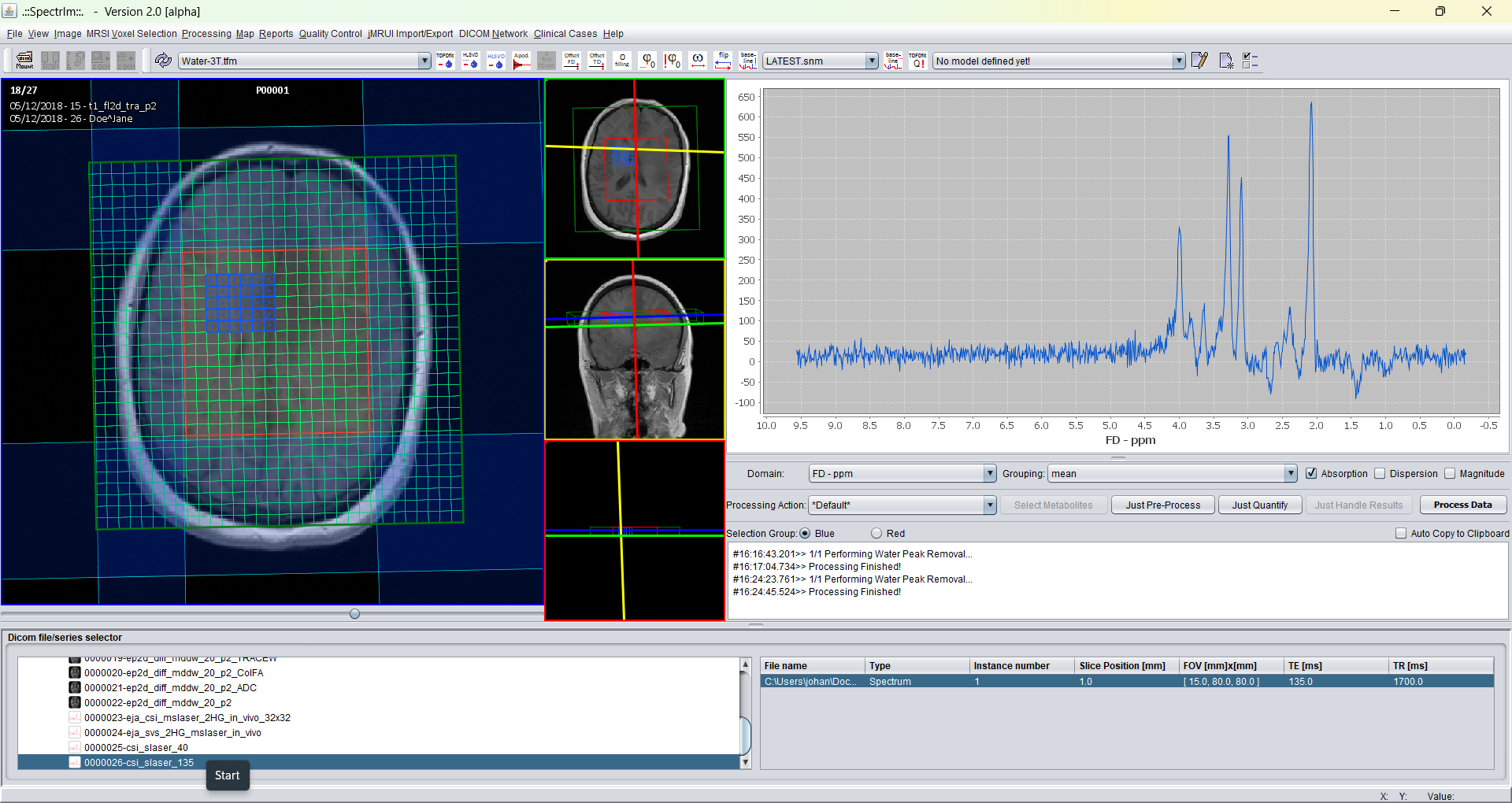
Before truncation from 1024 time domain points the sum spectrum of the selected volume displayed in blue is is the following:
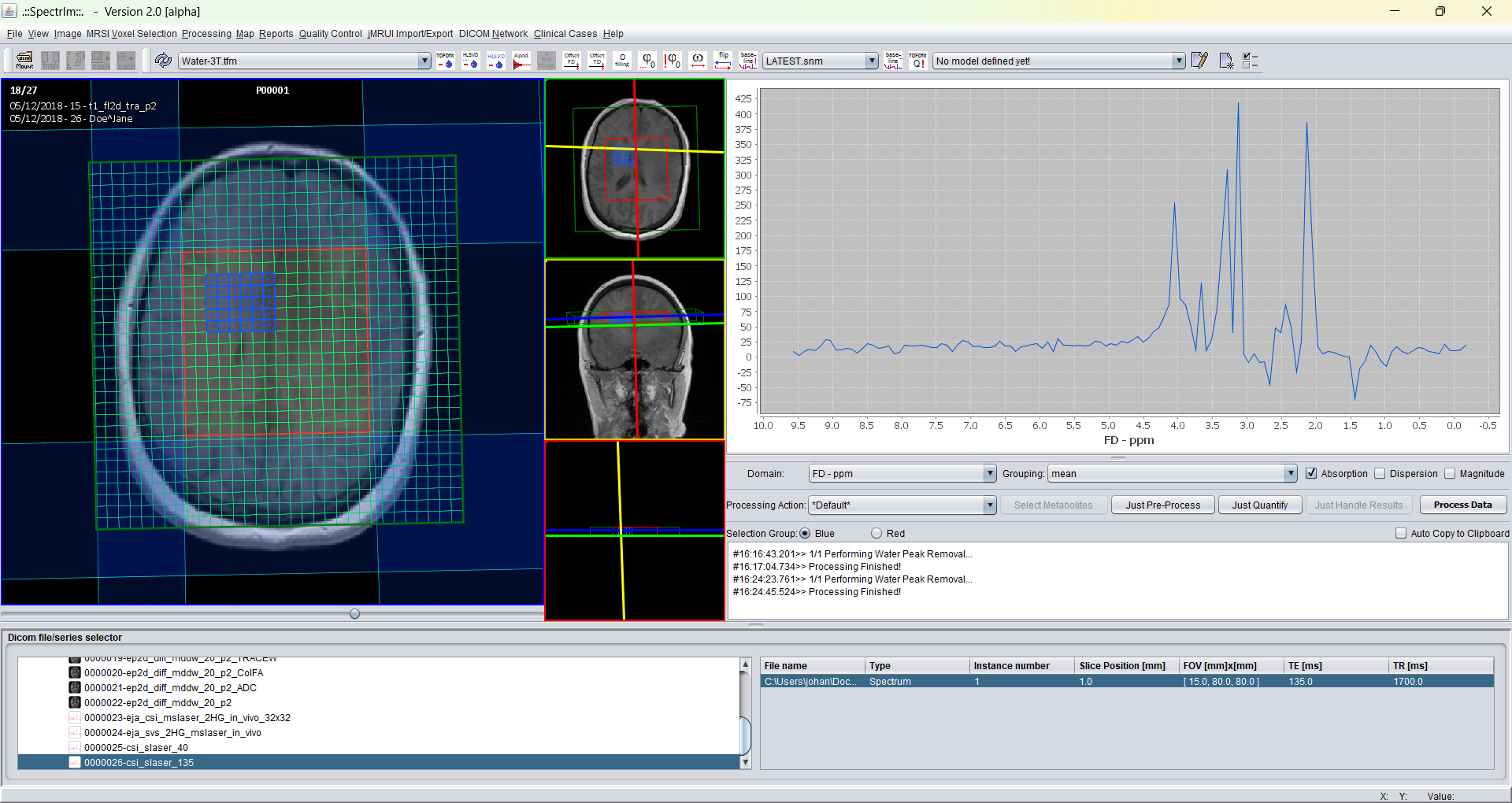
After truncation from 1024 to 128 time domain points the sum spectrum of the selected volume displayed in blue is is the following:
spectrIm-QMRS -> Processing -> Offset correction in Frequency Domain ..
After selection of the FD offset correction menu item the following popup window appears:
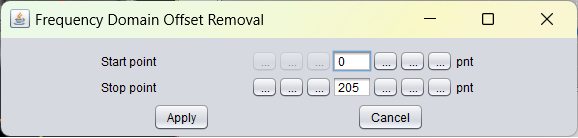
The graphical user interface enables the user set the two required parameter values to be used to perform a offset correction in FD with. The user needs to define a start point and a stop point in the spectra. Between these two values the mean for each individual spectrum is computed, and this mean is subtracted from each spectrum.
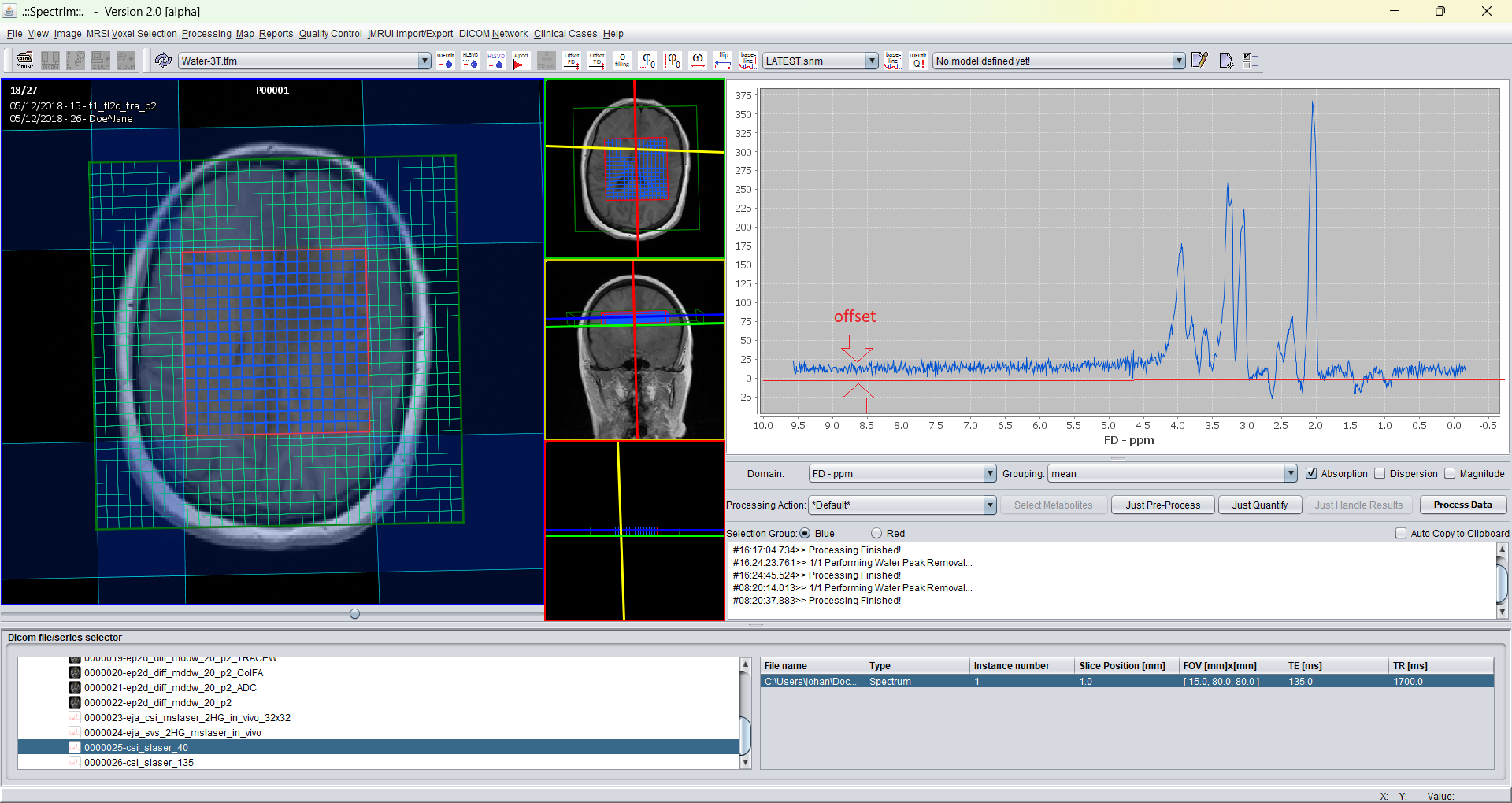
For 1H MRS(I) the default settings can often be used. After pressing the "Apply button" the spectrum looks the following:
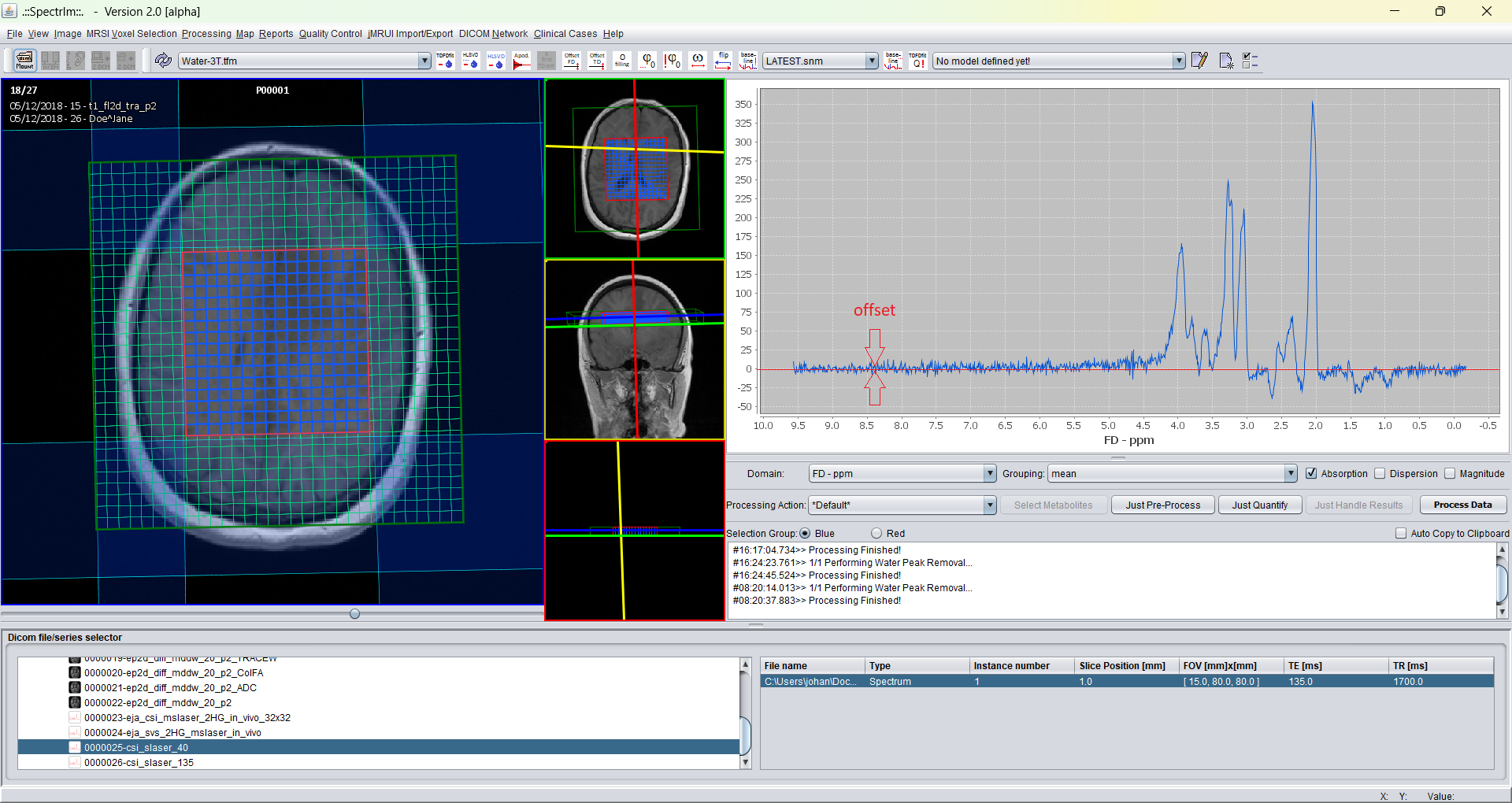
spectrIm-QMRS -> Processing -> Offset correction in Time Domain ....
After selection of the TD offset correction menu item the following popup window appears:
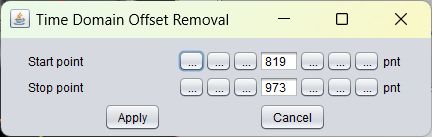
In contrast to FD offset removal, where a noisy part of the spectrum is selected at the begiing of the signal array, in TD offset removal the defaults in point number units are set at the end of the signal. In novel scanner systems however, TD-offset correction needs hardly ever to be used.
spectrIm-QMRS -> Processing -> Apodization..
Opens a graphical user interface that enables the user to set the type of apodization filter (Gaussian or Lorentzian) as well as the the line broadening factor in Hertz units. The graphical user interface is the following:
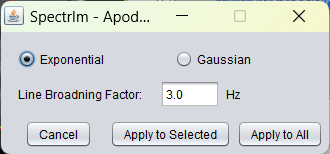
Example of the spectrIm-QMRS interface before apodization:
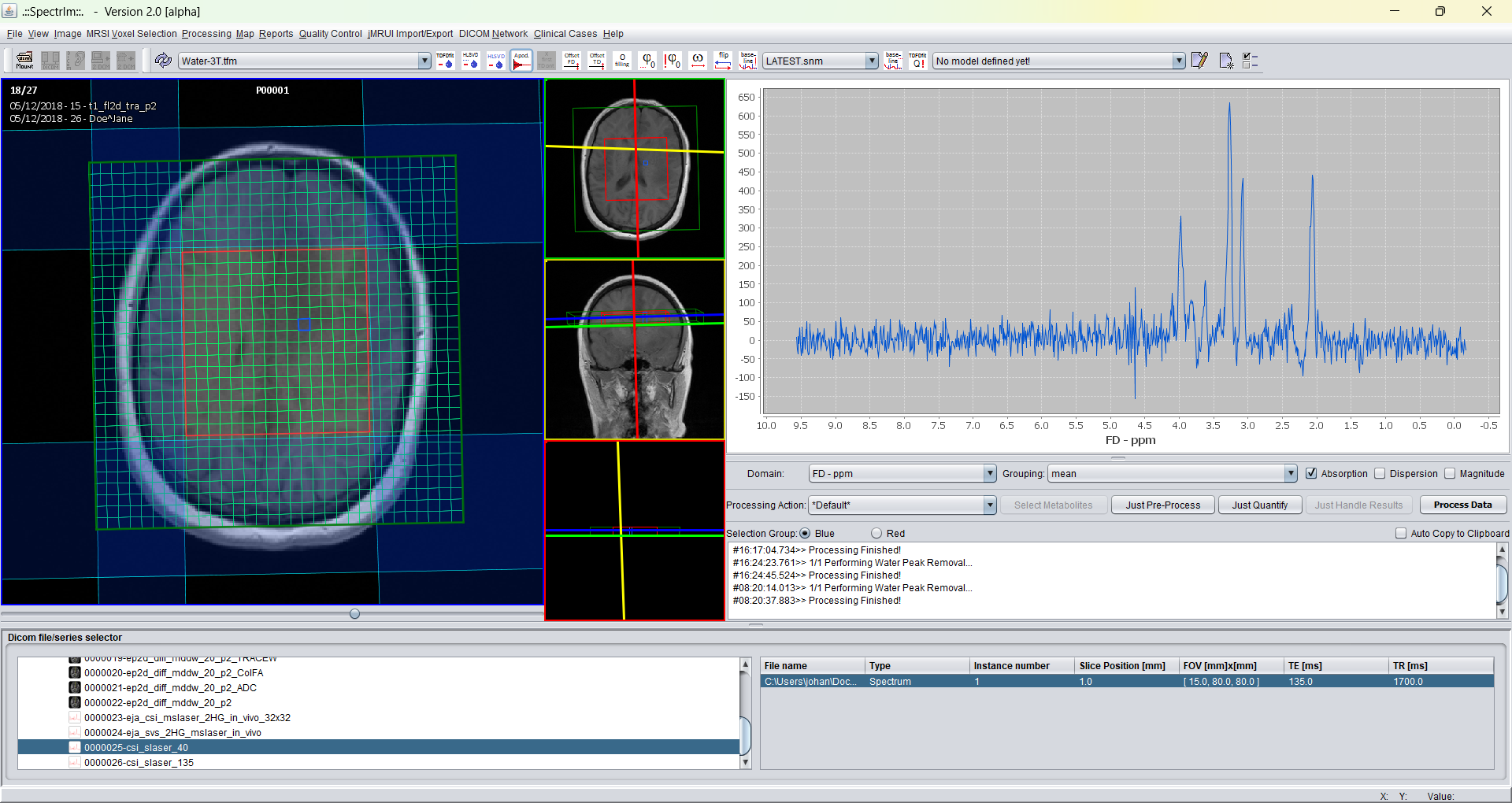
Example of the spectrIm-QMRS interface after apodization with a 3 Hz Lorentzian apodization filter. The effect of apodization is an optical better spectral SNR. The price to pay for this better SNR is a poorer spectral resolution. Apodization is not necessary before performing spectral fitting, due to the fact that it has only a cosmetic effect.
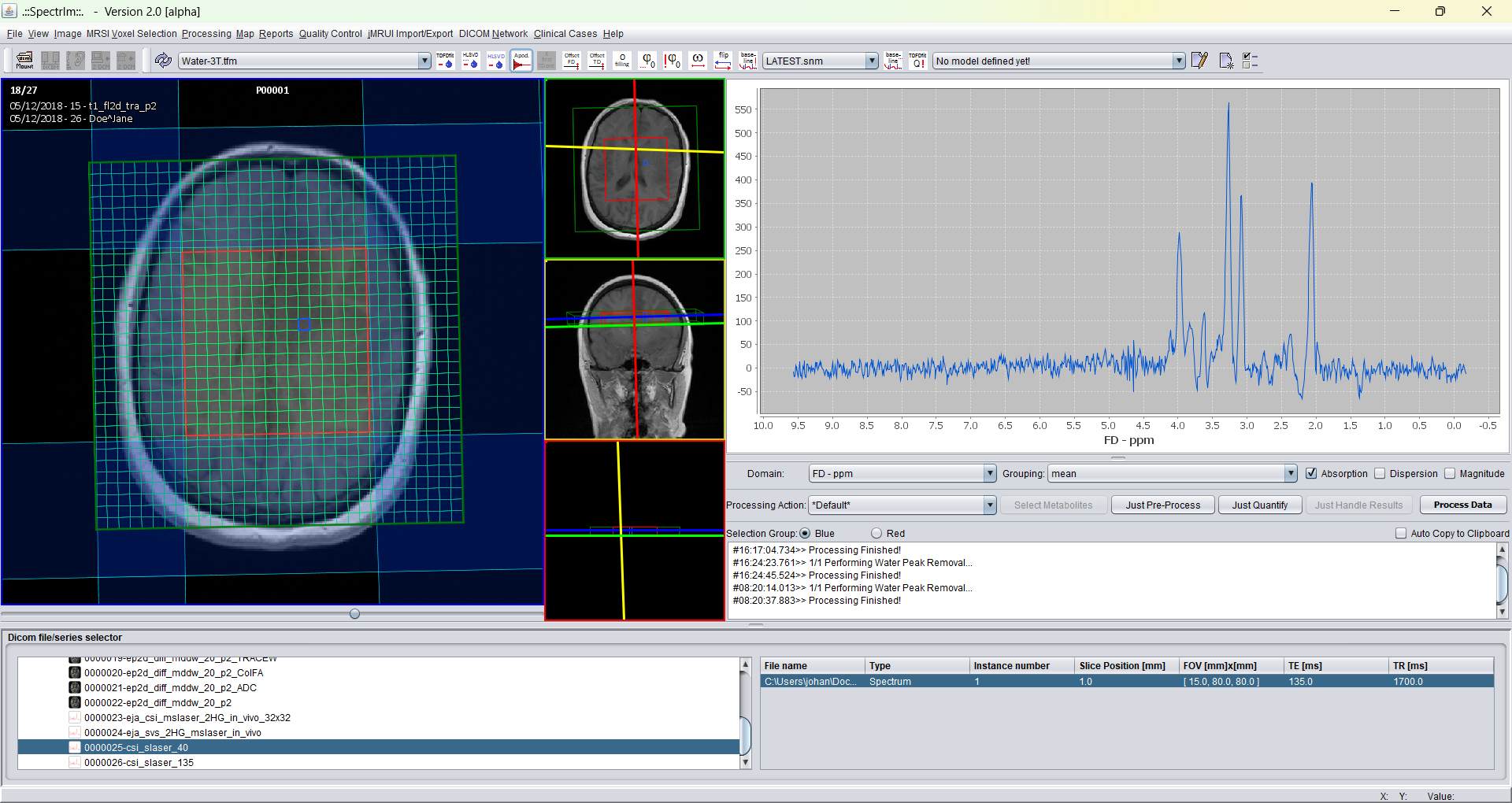
spectrIm-QMRS -> Correct Frequency Shift..
Opens a graphical user interface that enables the user correct the frequency shift for each spectral voxel inmdividually. The user can select the reference resonance lines to be used to perform the frequency shift upon. Multiple sections are allowed. Note that the selected reference line(s) must be present in the spectra of all sected voxels. For instance, selecting NAA as reference line in a high grade glioma may possibly not optimal within the tumor.
Defaults values are available for 1H MRS of the brain. The GUI looks the following:
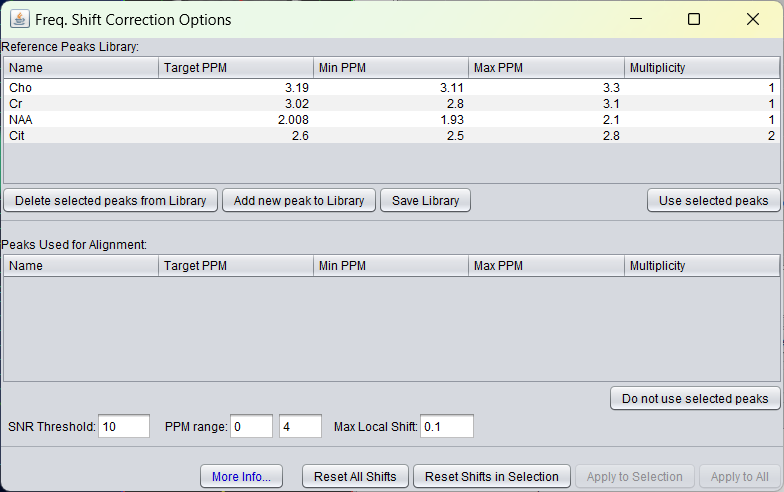
Again, the user has to select on which resonances the frequency has to be used to optimize the frequency shift. The list shows default values for NAA, Cho, Cr and Cit, and assumes 1H-MRS. The user can define more default metabolites, which can be stored and used in future use of spectrIm-QMRS. If the user selects NAA from the top list, this metabolite appears in the lower list. The GUI looks like following:
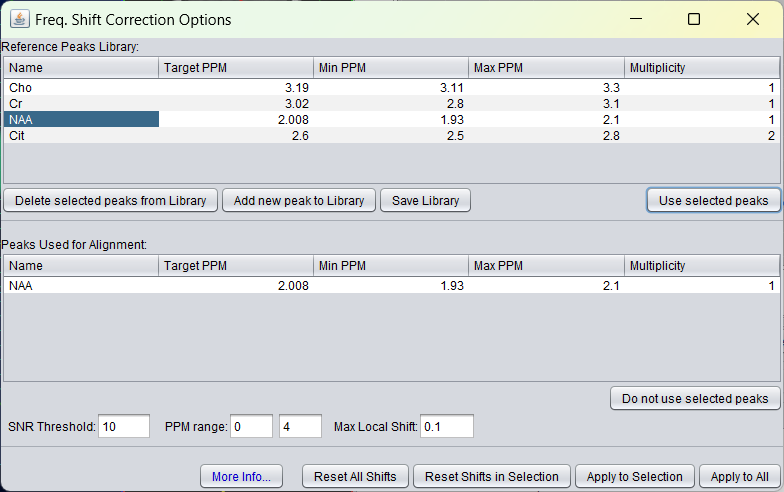
Before the application of frequency shift alignment on NAA the sum spectrum of a 2D MRSI looks like follows:
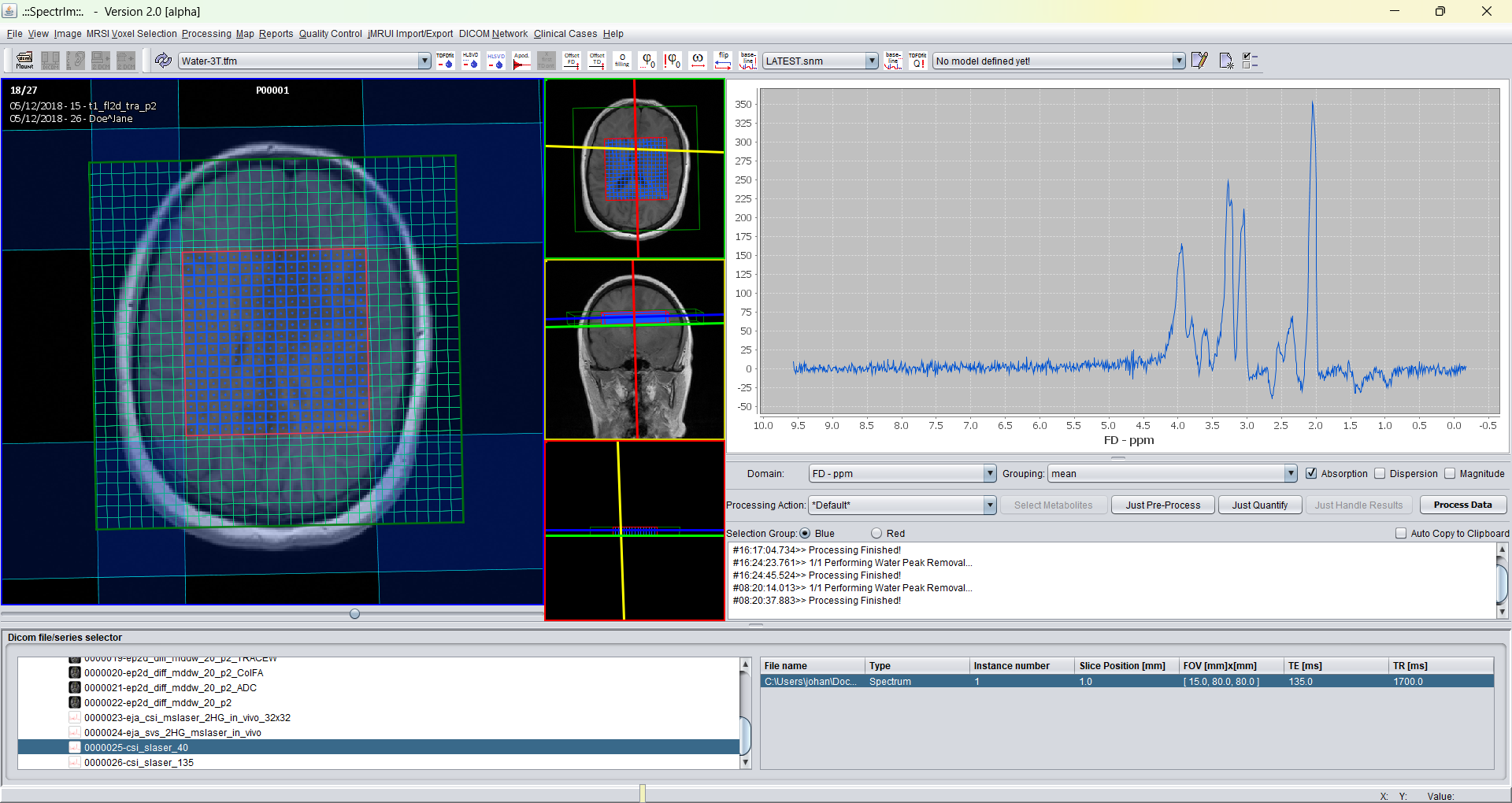
After the application of frequency shift alignment using the NAA peak as reference peak the sum spectrum of a 2D MRSI looks like follows:
The alignment operation is launched by pressing either "Apply to selection" or "Apply to all" button. Please note the dramatic increase in spectral resolution of the sum spectra after frequency alignment. Frequency alignment is highly recommended to perform before the spectral fitting is performed. The reason for this is that spectral fitting performs best, if the starting values are already as close as possible to the optimum values.
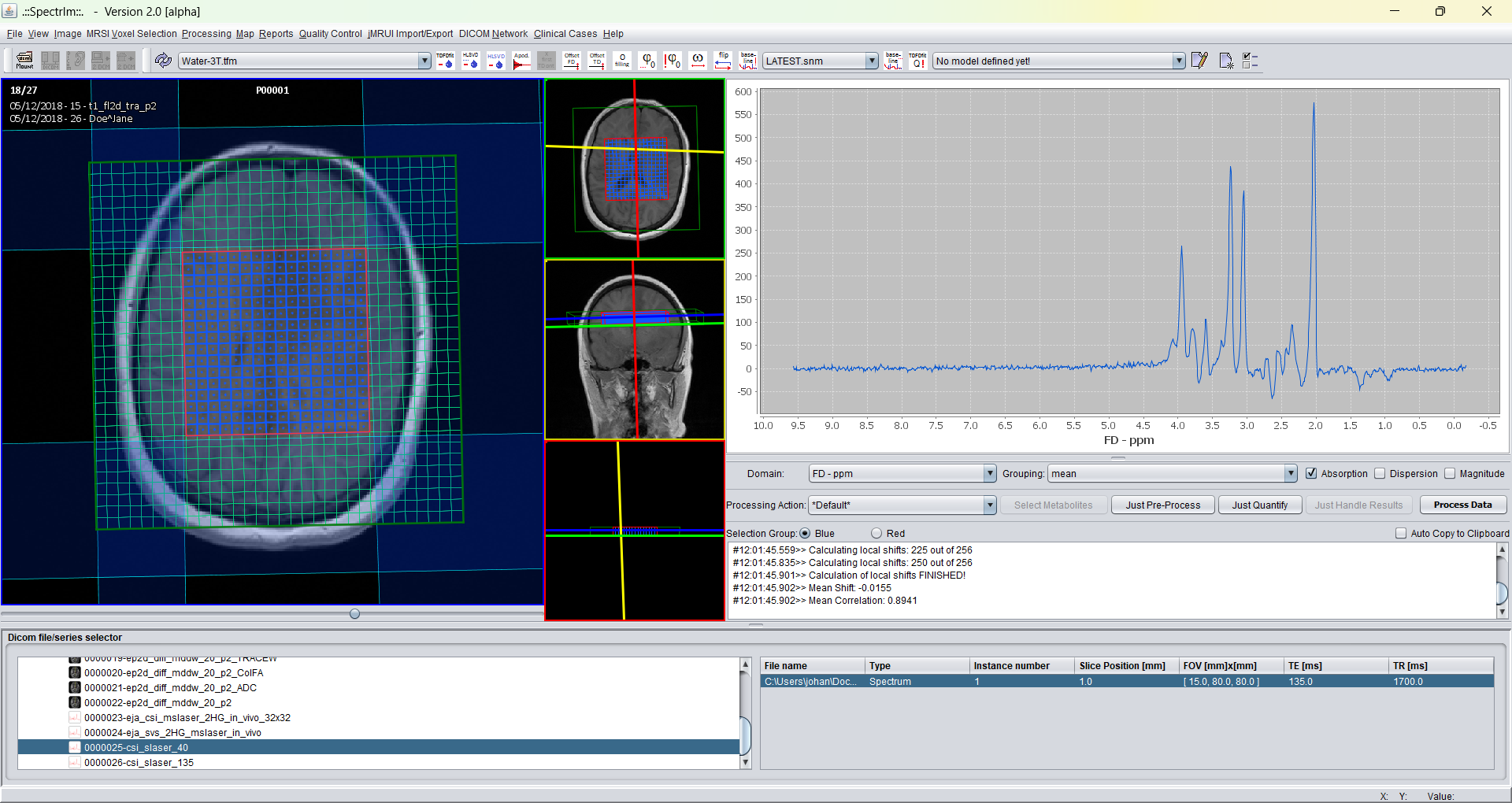
spectrIm-QMRS -> Processing -> Auto phasing..
Opens a graphical user interface that enables the user to perform automatic zero order phasing of the selected spectra. The automatic phasing
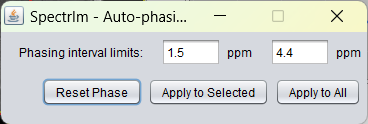
The user has to define a minimum offset frequency value in ppm and a maximum offset value in ppm. In this ppm value range the algorithm will maximimize the real part of spectrum integral value. Please note that this will give satisfactory results, if the spectrum is not dominated by residual water or lipid contributions which overlap with the spectral frequency range. For 1H-brain MRS(I) the set defaults values are working fine if above mentioned condition is met. Example: the sum spectrum looks the following before automatic phasing:
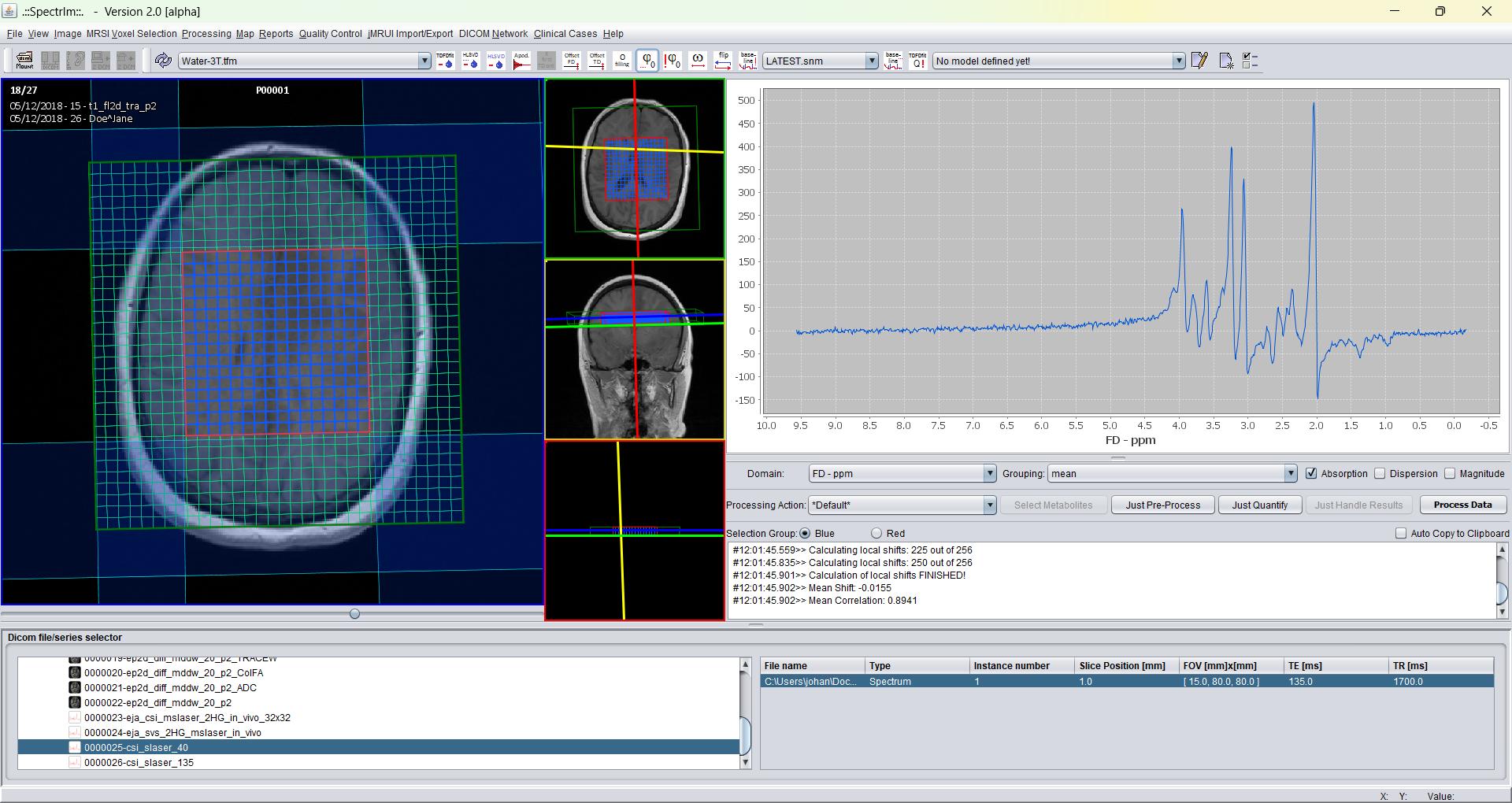
And after application of automatic zero order phasing:
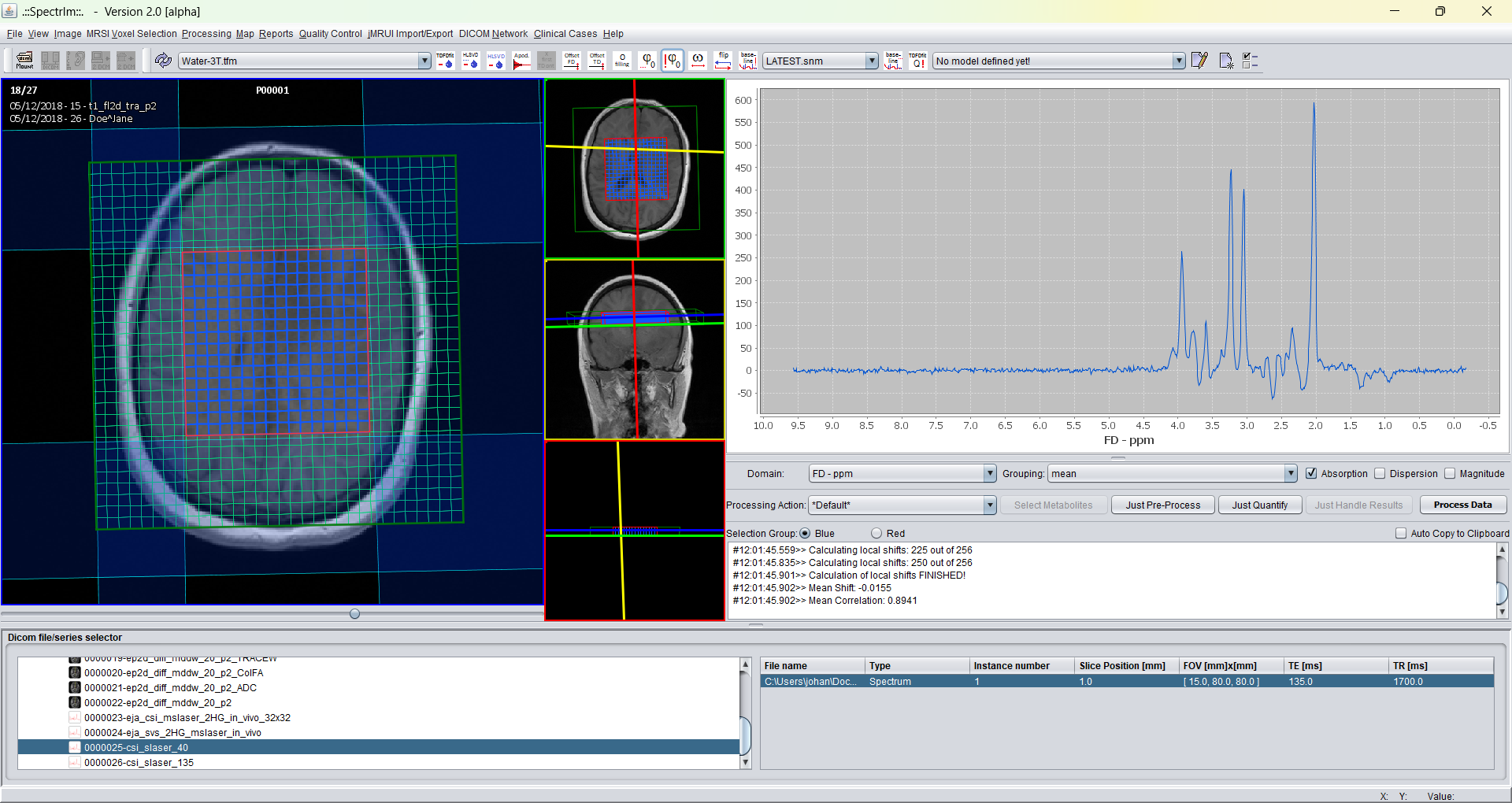
Note the increasing phase deviation going from the left to the right in the spectrum. This is due to the misset linear first phase over the spectrum of 360 degrees. Also note the broad baseline component which becomes visible.
spectrIm-QMRS -> Processing -> Manual Phasing..
Opens a graphical user interface that enables the user to manually zero and first order phase the selected spectra.
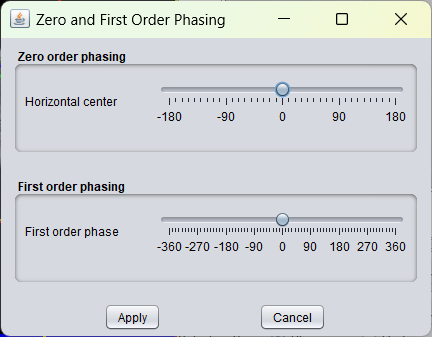
By moving the above slider, the zero order phase correction value for the selected spectra can be set. By moving the lower slider, the first order phase correction values can be set.
Drawback of the current implementation is that the first order can only be set between -360 and + 360 degrees. In a future version the first order phase range will be defined broader.
To illustrate first order phasing, the GUI below displays the application of a first order phasing of + 360 degrees on an initially well phased spectrum.
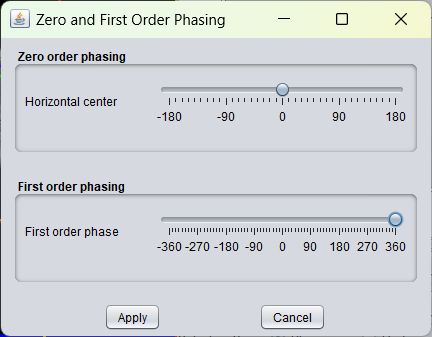
Below see the effect this first order phasing about + 360 degrees on an initially well-phased spectrum.
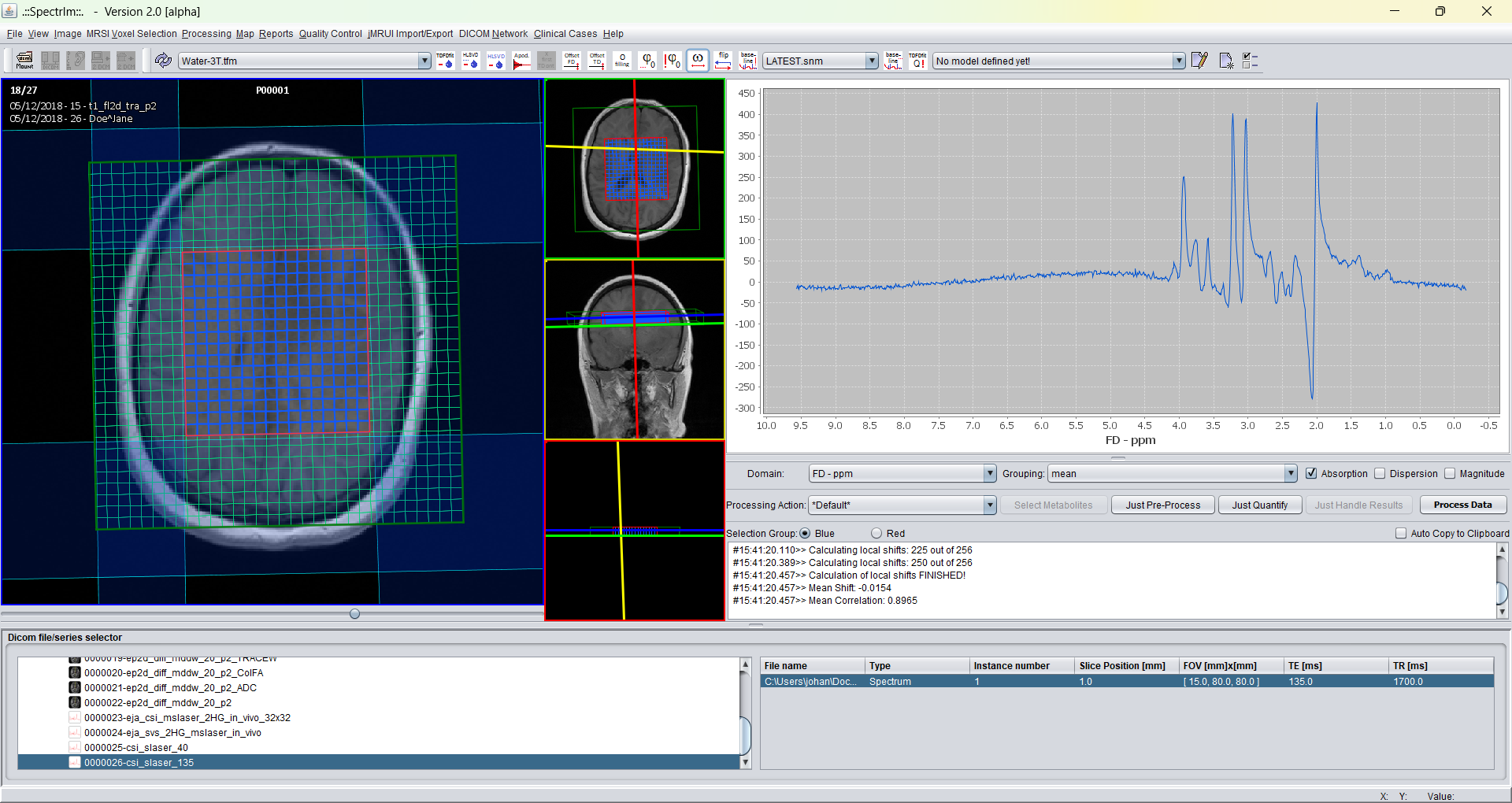
spectrIm-QMRS -> Processing -> Flip Spectra (complex conjugation)
Invoking the flip spectra menu item flips the spectra horizontally: high frequencies become low frequencies and vice versa. Twice application restores the original state again.
The original spectra:
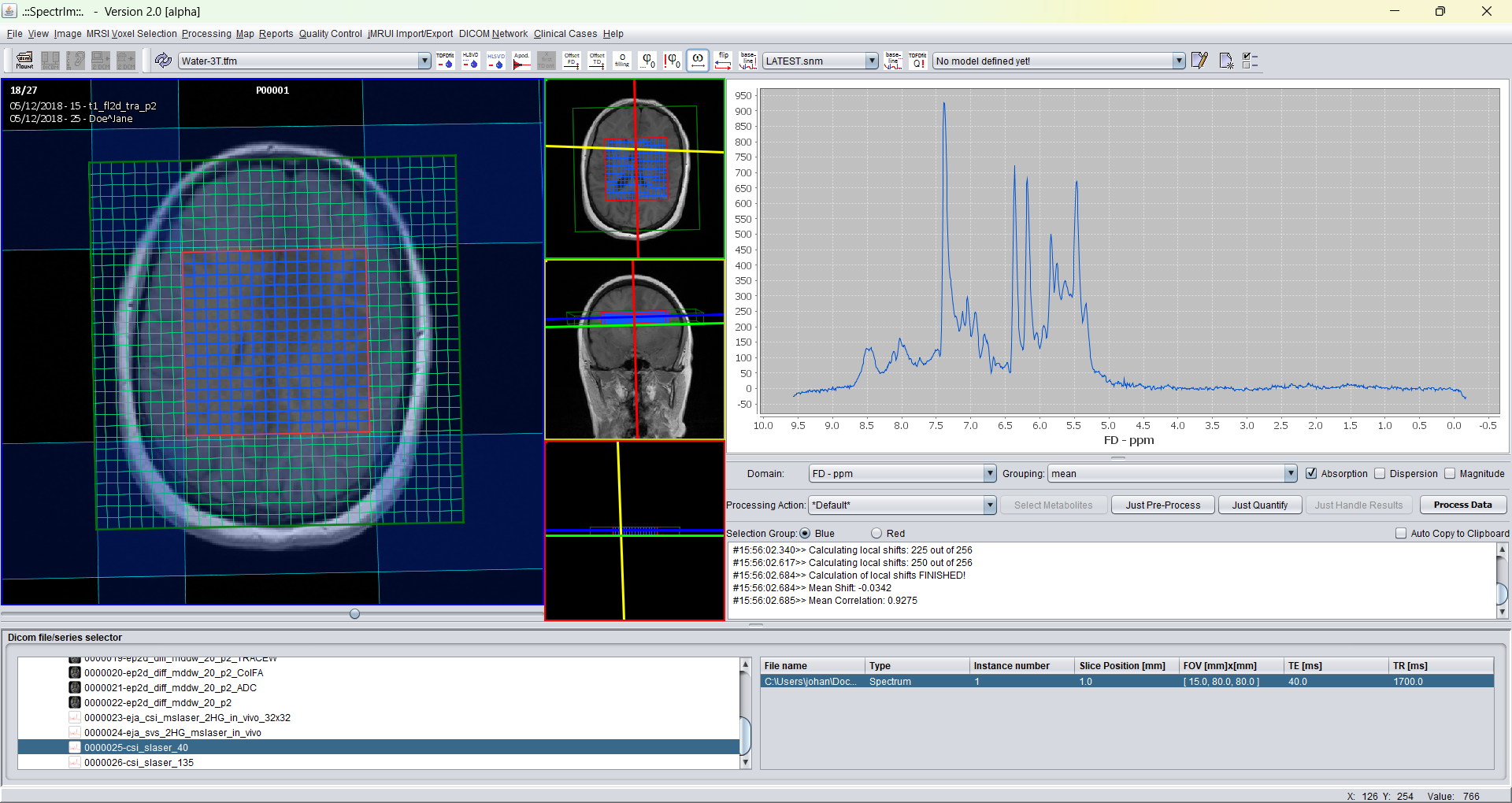
And after flipping the sum spectra looks the following way:
spectrIm-QMRS -> Processing -> Write Quantification results
If the quantification of the data using TDFDFit has taken place, the user can write the results of the quantification to a file in the file system.
If spectrIm-QMRS is launched as a stand-alone application, that means NOT as plugin of 32-bits jMRUI, calling this function has no effect.
spectrIm-QMRS -> Processing -> Print Quantitation for Selected Voxels
If the quantification of the data using jMRUI had taken place, the user can print the results of the quantification from the jMRUI-kernal.
If spectrIm-QMRS is launched as a stand-alone application, that means NOT as plugin of 32-bits jMRUI, calling this function has no effect.
spectrIm-QMRS -> Processing -> Print Quantitation Results
If the quantification of the data using jMRUI had taken place, the user can print the results of the quantification to jMRUI-kernal.
If spectrIm-QMRS is launched as a stand-alone application, that means NOT as plugin of 32-bits jMRUI, calling this function has no effect.
spectrIm-QMRS -> Processing -> Create new TDFDFit model..
Selecting "Create new TDFDFIT model.." opens a separate window in which a new TDFDFit model can be created. TDFDFit is a non linear least squares fitting algorithm that uses, alike LCModel and Quest, a pre-calculated metabollite basis sets to perform the fitting with. These basis sets can be used in combination with a number Voigt-lines. You may find more information on TDFDFit here .
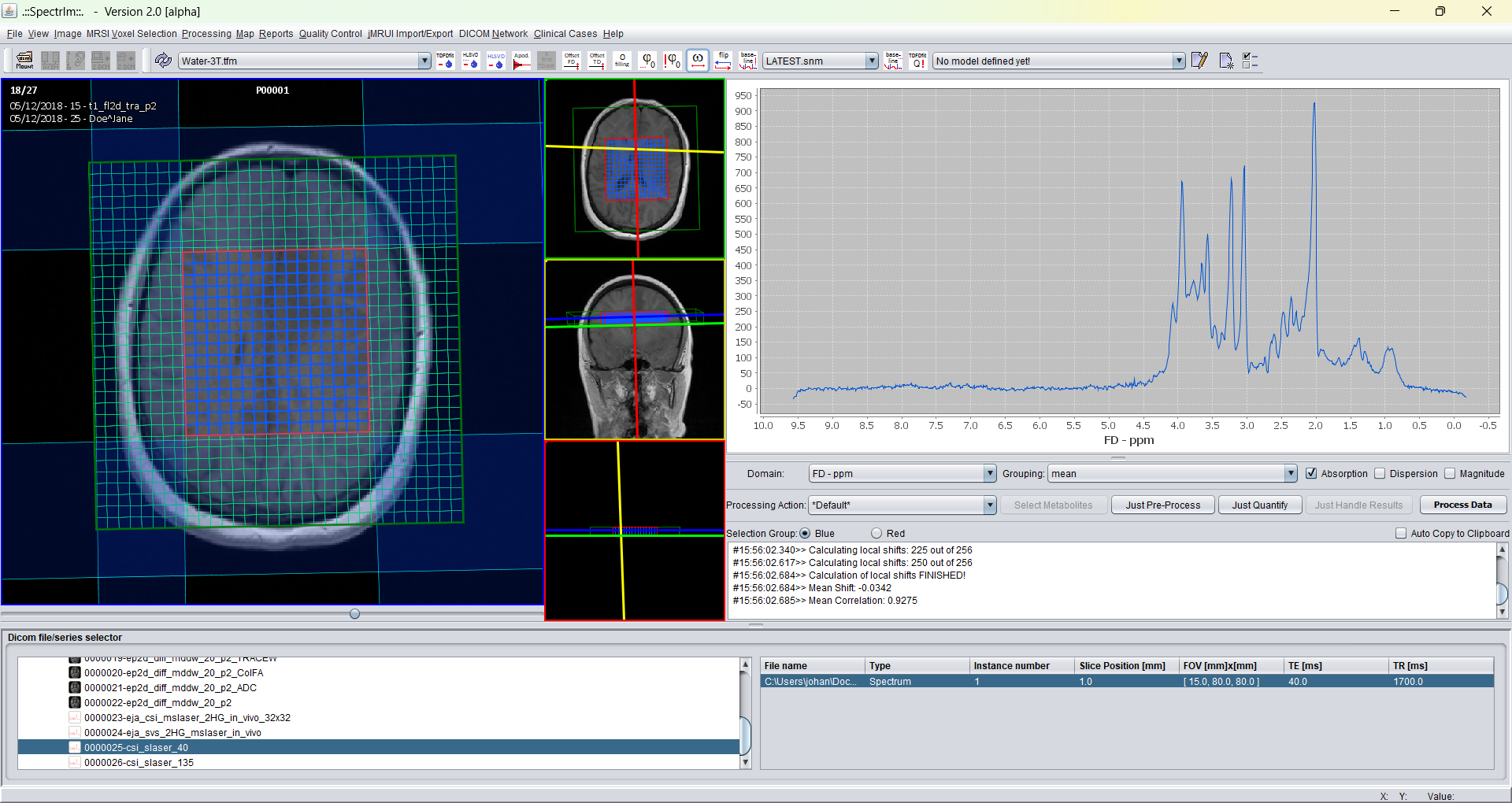
Prerequisit for being able to successfully define a TDFDFit prior knowledge model is that a spectroscopic dataset have been loaded, and at least one spectrum is selected. Assume that the loaded dataset looks like following:
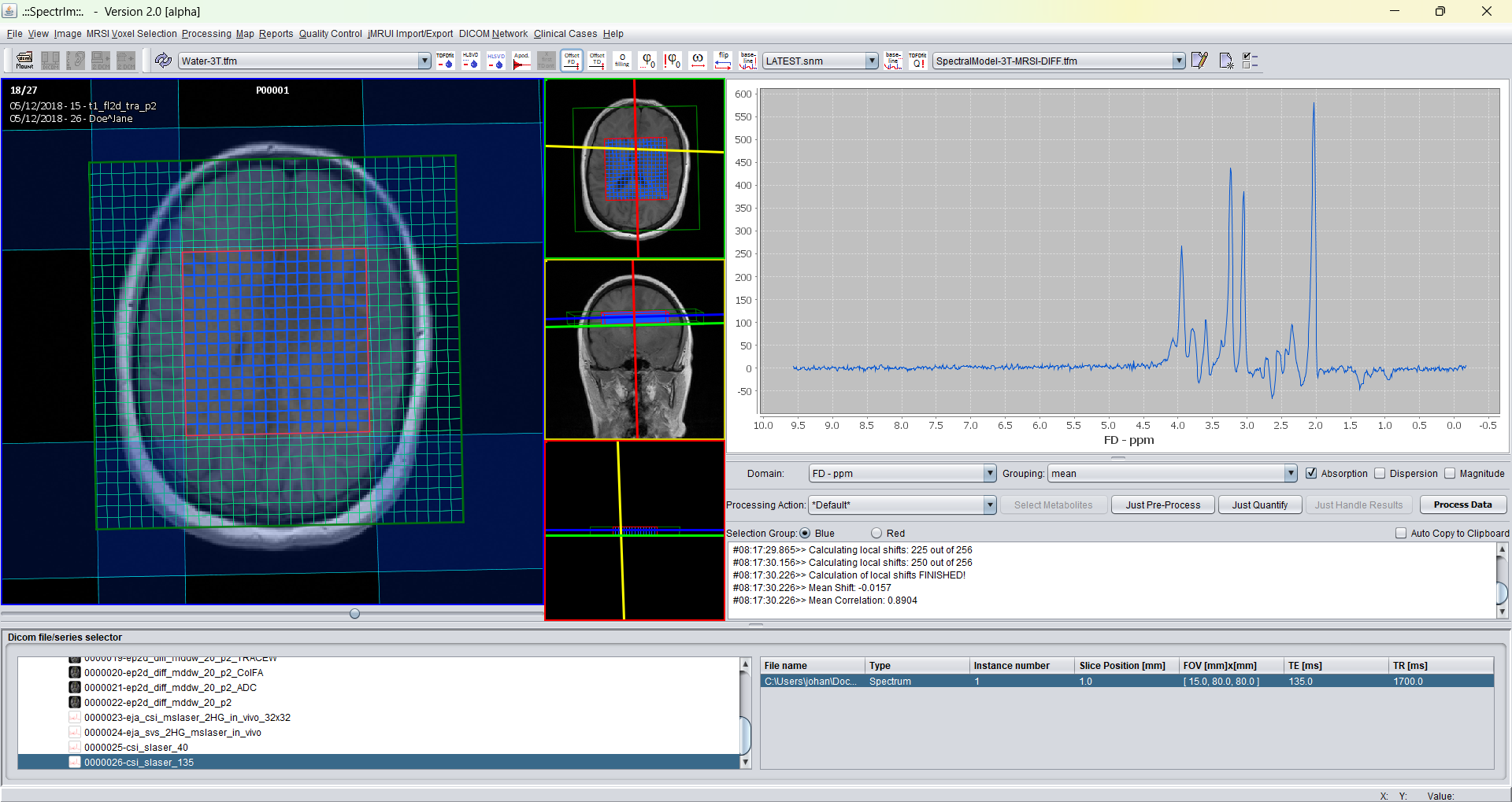
After invoking the "Create new TDFDFit model.." two windows popup as follows:
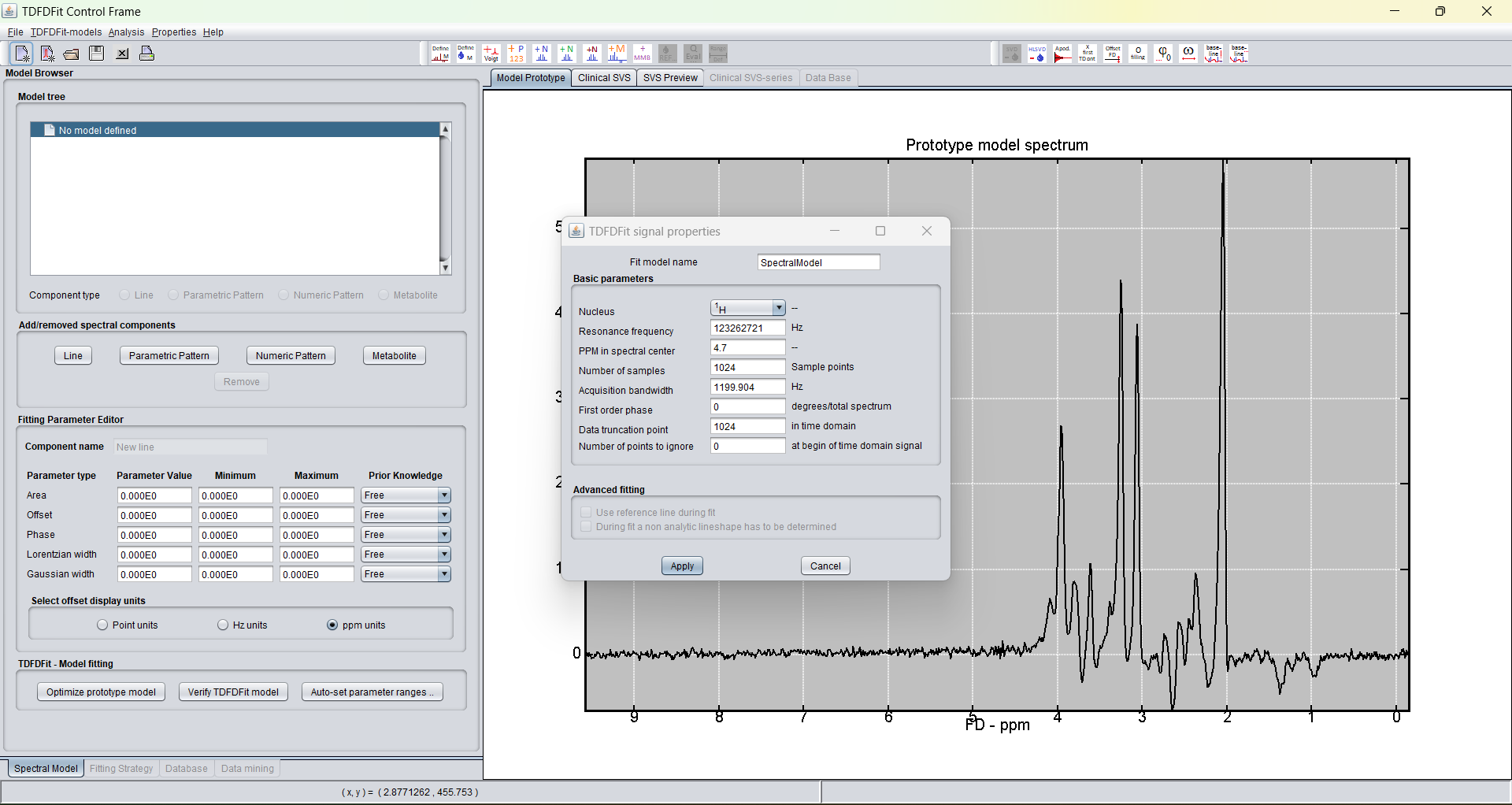
The big window is the "TDFDFit Control" which enables to user to define prior-knowlegde based fitting models. Please consult the TDFDFit Control web pages.
The small popup window in front of the larger "TDFDFit Control" window gives an overview of the signal properties of the loaded spectra. The user has the possibility to set some parameters that will be used in the further modelling process of the spectrum which displayed.
The displayed spectrum in the "TDFDFit Control" is the so-called the prototype spectrum. The prototype spectrum should be selected such, that the (prototype) TDFDFit model is close to the average quality of the spectra one wants to fit with in in future.
spectrIm-QMRS -> Processing -> Outlier correction ..
Not available in this release of the software.
spectrIm-QMRS -> Processing -> Quantify spectra (TDFDFit) ..
Under construction!
spectrIm-QMRS -> Processing -> Automatic store of TDFDFit Results after fitting ..
Under construction!
spectrIm-QMRS -> Processing -> Print TDFDFit results to screen ..
Under construction!
spectrIm-QMRS -> Processing -> Write TDFDFit results to file ..
Under construction!
spectrIm-QMRS -> Processing -> Load quantification results from file system ..
Loads previously stored quantification information, if available. If the toggle to store automatically store TDFDFit results after fit, the data is persistendly available in the file system.
spectrIm-QMRS -> Processing -> Process all spectroscopy series (mounted) ..
Under construction!
spectrIm-QMRS -> Processing -> Process all spectroscopy series in folders ..
Under construction
spectrIm-QMRS -> Processing -> Read matching preprocessed data ..
Reads the preprocessed data to the file system. -- Under construction
spectrIm-QMRS -> Processing -> Write matching preprocessed data ..
Writes the preprocessed data to the file system. -- Under construction