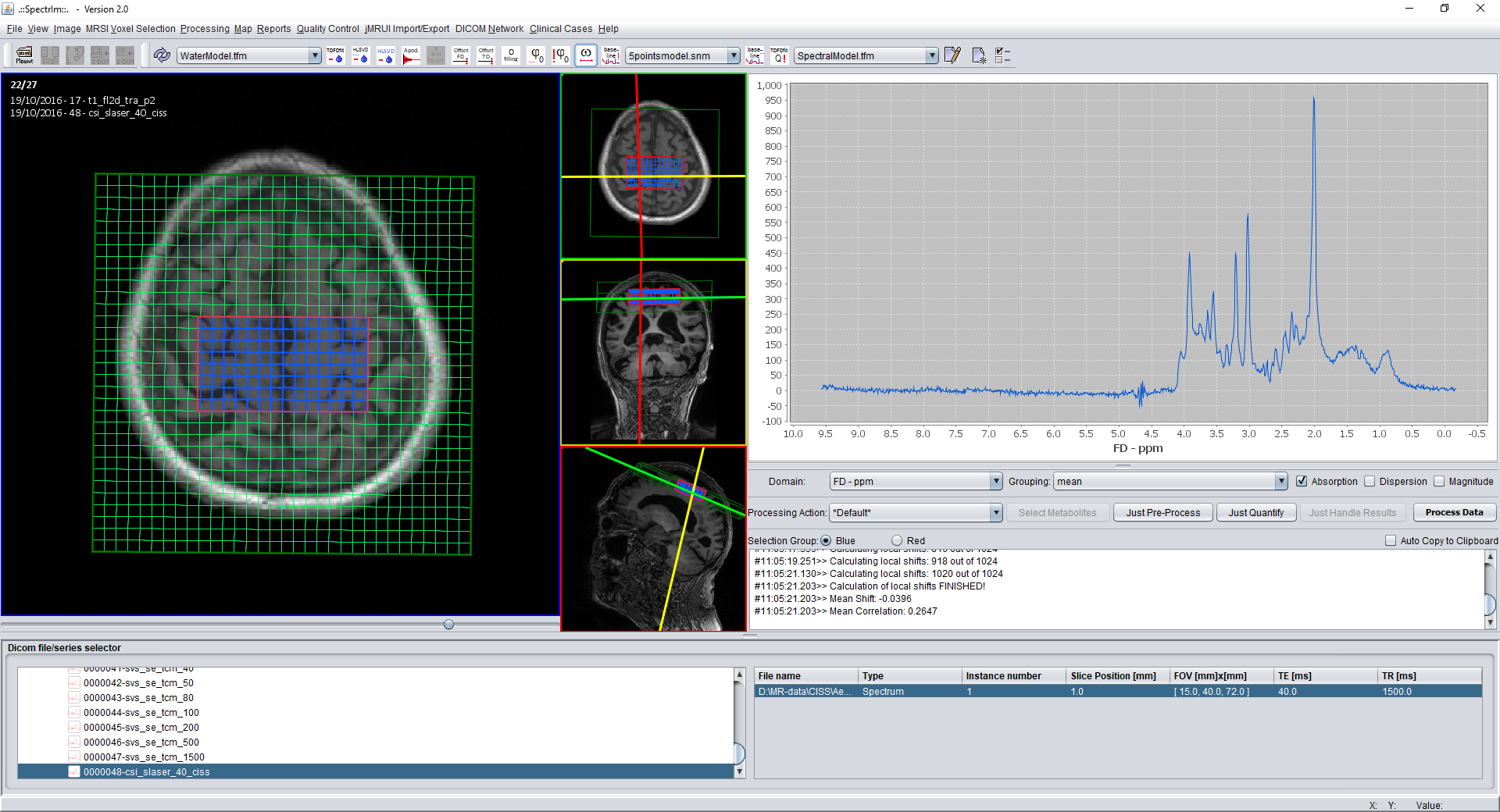
spectrIm-QMRS -> MRSI Voxel Selection menu-> Select all voxels
Select all voxels (also accessable by pressing simultaniously 'Ctrl + A' on the keyboard.
The effect on a loaded 2D-MRSI dataset will be the following:

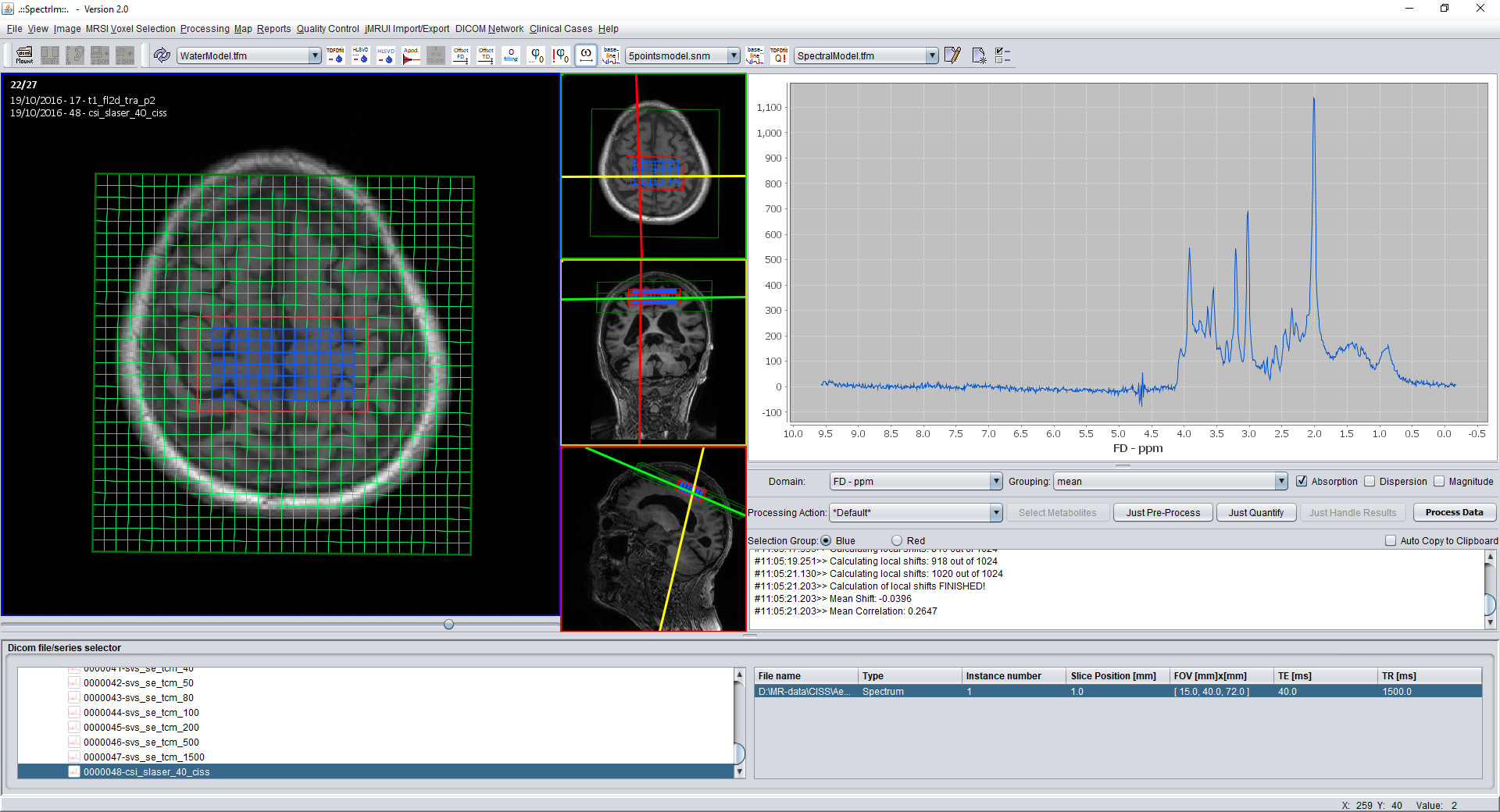
spectrIm-QMRS -> MRSI Voxel Selection menu-> Select excited voxels
Select excited voxels only (also accessable by pressing simultaniously 'Ctrl + E' on the keyboard.
The effect on a loaded 2D-MRSI dataset will be the following:
spectrIm-QMRS -> MRSI Voxel Selection menu-> Select inner excited voxels
Select the inner excited voxels only (also accessable by pressing simultaniously 'Ctrl + Shift + E' on the keyboard.
The effect on a loaded 2D-MRSI dataset will be the following:
spectrIm-QMRS -> MRSI Voxel Selection menu-> Select inner excited voxels
Select the inner excited voxels only (also accessable by pressing simultaniously 'Ctrl + Shift + E' on the keyboard.
The effect on a loaded 2D-MRSI dataset will be the following:

spectrIm-QMRS -> MRSI Voxel Selection menu-> Spectral feature based selection
This functionality enables MRSI voxel selection based upon spectral features computed from the spectra/signals (also accessable by pressing simultaniously 'Ctrl + Shift + F' on the keyboard.
The effect on a loaded 2D-MRSI dataset will be that a popup GUI appears enabling the user to define feature based selection criteria:
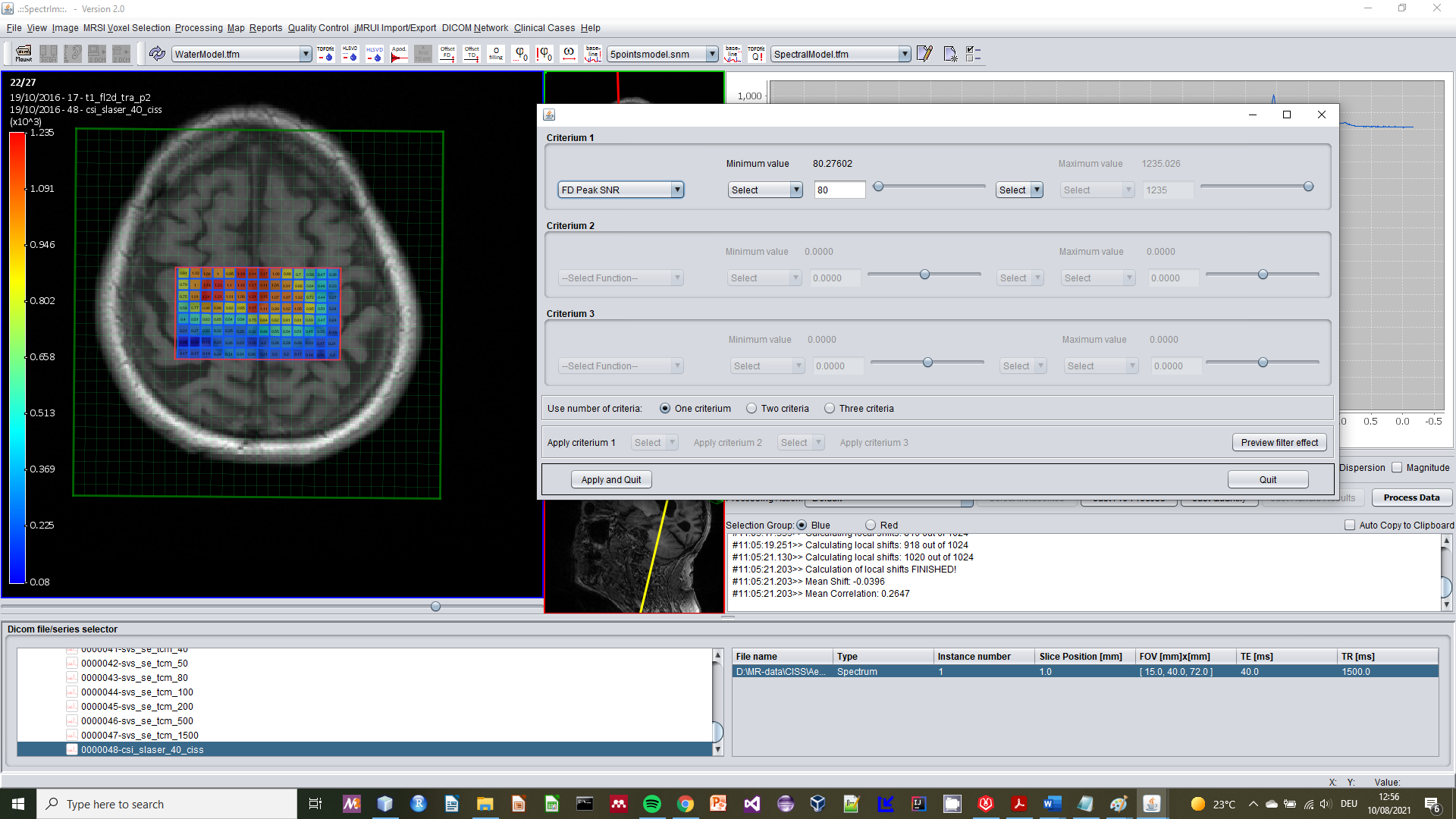
The current status of the functionality is "experimental" and is NOT recommended for use yet!
spectrIm-QMRS -> MRSI Voxel Selection menu-> Freehand selection
This functionality enables MRSI voxel selection by the user by drawing a contour around all voxels which should be selected. It can also be envoked by pressing 'Ctrl + D' on the keyboard.
The current status of the functionality is "experimental" and is NOT recommended for use yet!
spectrIm-QMRS -> MRSI Voxel Selection menu-> Good quality only
This functionality enables MRSI voxel selection by selecting only those voxels for which sufficient spectral quality is reached. This functionality can only be used if classifiers are available that define what 'Ctrl + Alt + Q' on the keyboard.
The current status of this functionality is "experimental" and is NOT recommended for use yet!
spectrIm-QMRS -> MRSI Voxel Selection menu-> Remove saturated voxels
This functionality enables MRSI voxel selection by removing all voxels which are in the pulse sequences' saturation bands. This functionality can also be accessed by pressing 'Alt + S' on the keyboard.
Example: Before envoking the functionalty:
Example: After envoking the functionalty:

The current status of the functionality is "experimental" and not generally applicable: the reason is that is it will work properly on Siemens spectroscopic data only.
spectrIm-QMRS -> MRSI Voxel Selection menu-> Change selection Group
This functionality enables MRSI voxel selection by removing all voxels which are in the pulsesequences' saturation bands. This functionality can also be accessed by pressing 'Space' on the keyboard.
Example: After selection of the menu item or the blue radio button group: selected voxels will be added to the blue voxel selection group:
Example: After selection of the menu item or the blue radio button group: selected voxels will be added to the blue voxel selection group:
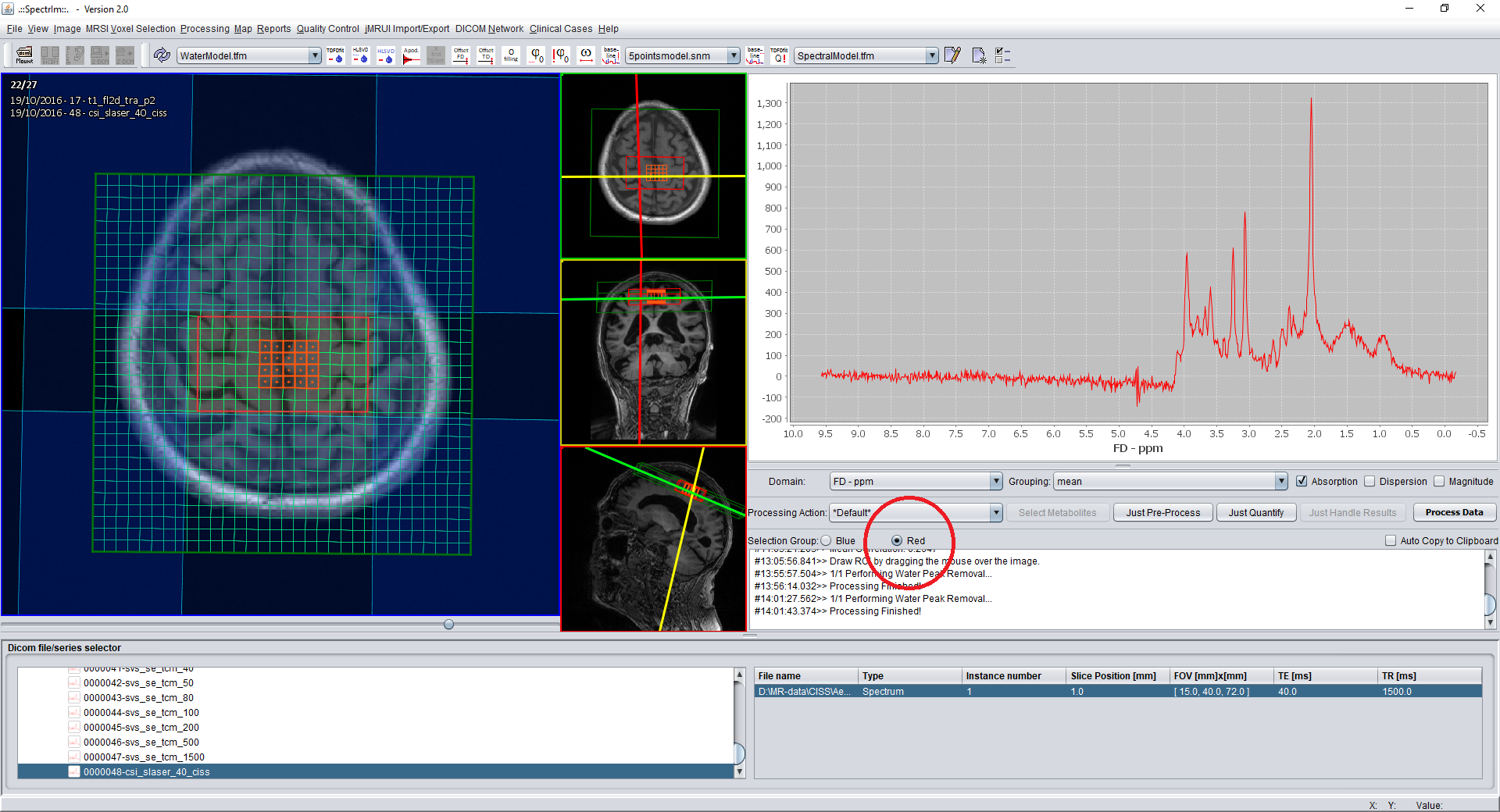
The current status of the functionality is "experimental" and not generally applicable: the reason is that is it will work properly on Siemens spectroscopic data only.
spectrIm-QMRS -> MRSI Voxel Selection menu-> Mirror Selection
This functionality enables already selected MRSI voxels of the currently selected (read or blue) selection group to be spatially mirrored and put into the complementary voxel group. This functionality can also be accessed by pressing 'Ctrl + Space' after at least one voxel has been selected. The example below tries to make graphically clear what is done exactly.
br>Example: The user selects the healthy tissue voxels on the lefthand side of the spectral grid: selected voxels will be added to the blue voxel selection group:
br>
img src = "images/006-spectrim-qmrs-mrsi-before-mirror-selection.png" width=920 height=512 alt = "User selected a region in healthy tissue of the brain to be added to the blue group." />
br>
br>Example: After selection of the menu item 'Mirror Selection' a new selection group is added (in this case red) which exactly mirrors the blue selection group. The mirror axis is the vertical line through the center of the excite volume.
br>
img src = "images/006-spectrim-qmrs-mrsi-after-mirror-selection.png" width=920 height=512 alt = "The 'mirror selection' adds a red selection group from the blue group by selecting " />
Te current status of the functionality is "experimental" and not generally applicable: the reason is that is it will work properly on Siemens spectroscopic data only.
/p>
spectrIm-QMRS -> MRSI Voxel Selection menu-> XYPlots of selection
This functionality enables the user to make a multi xy-plots of the selected spectral voxels. For instance this selection:

results in following grid of XY-plots:
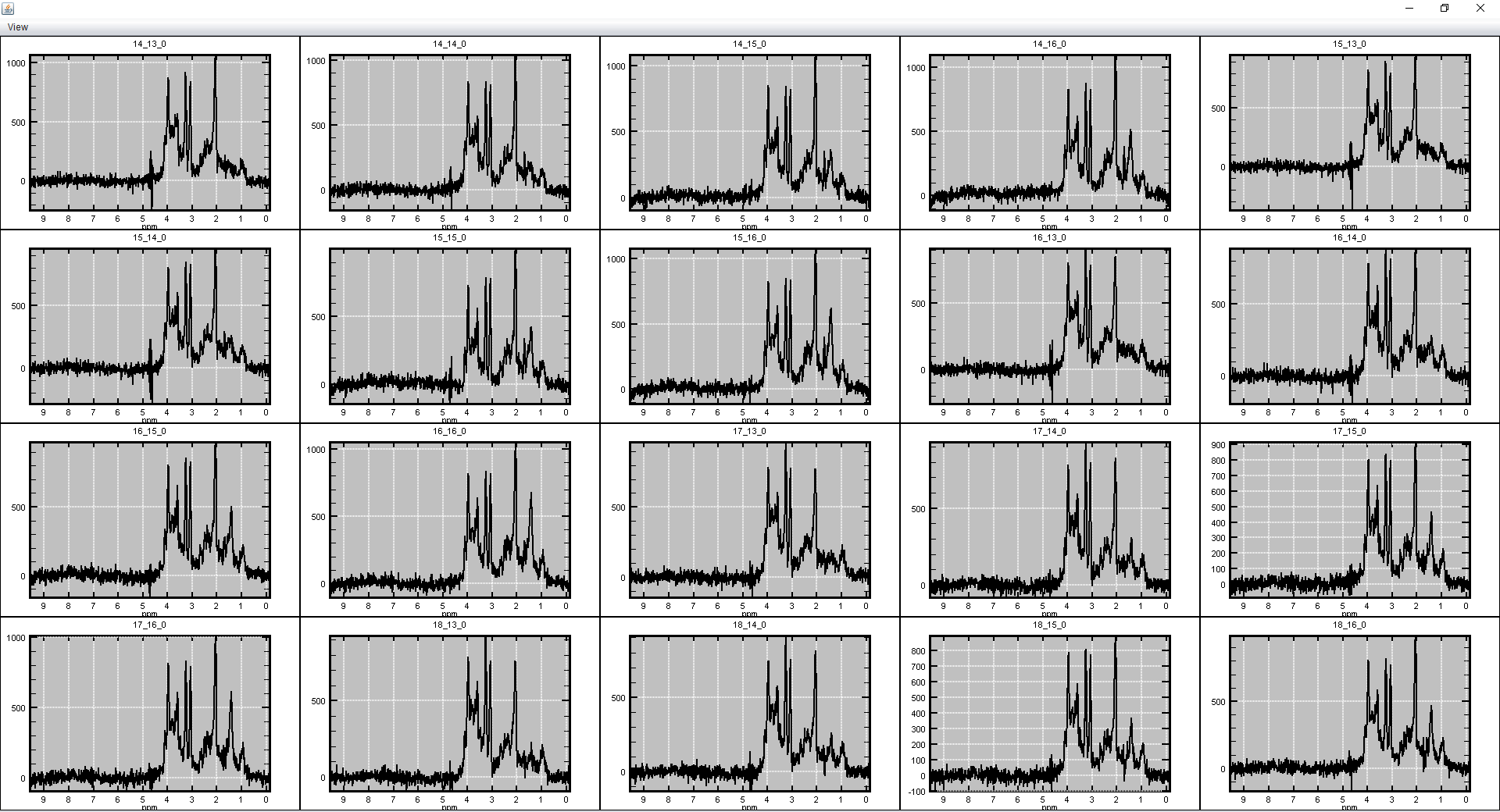
The user can sent the XY plot grid to a printer, or store the XY plotgrid as a png-formatted file.
spectrIm-QMRS -> MRSI Voxel Selection menu-> Save selection as preset ..
This functionality enables the user to the current selection persistent, that means the selected voxels are stored in the file-system and can be loaded lateron. This has the advantage that selections can be used multiple times in a reprocible way.

As a result the preset are loadable for the user at a later time.
spectrIm-QMRS -> MRSI Voxel Selection menu-> Add to current selection
After the user selected the second preset via the menu, this preset gets loaded and, if the 'Add to Current Selection' checkbox menu item is selected, is added to the already defined selected voxels:
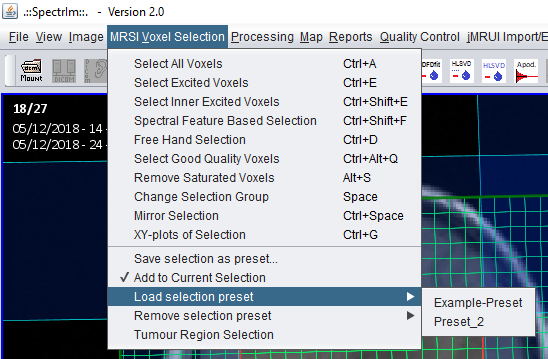
spectrIm-QMRS -> MRSI Voxel Selection menu-> Load selection preset -> ...
After the user selected the second preset via the menu, this preset gets loaded and, if the 'Add to current selections' checkbox menu item is selected, is added to the already defined selected voxels as can be seen in the example below:

spectrIm-QMRS -> MRSI Voxel Selection menu-> Remove selection preset -> ...
After removal of the second preset via the menu (same example as above) the GUI of spectrIm-QMRS looks as follows:
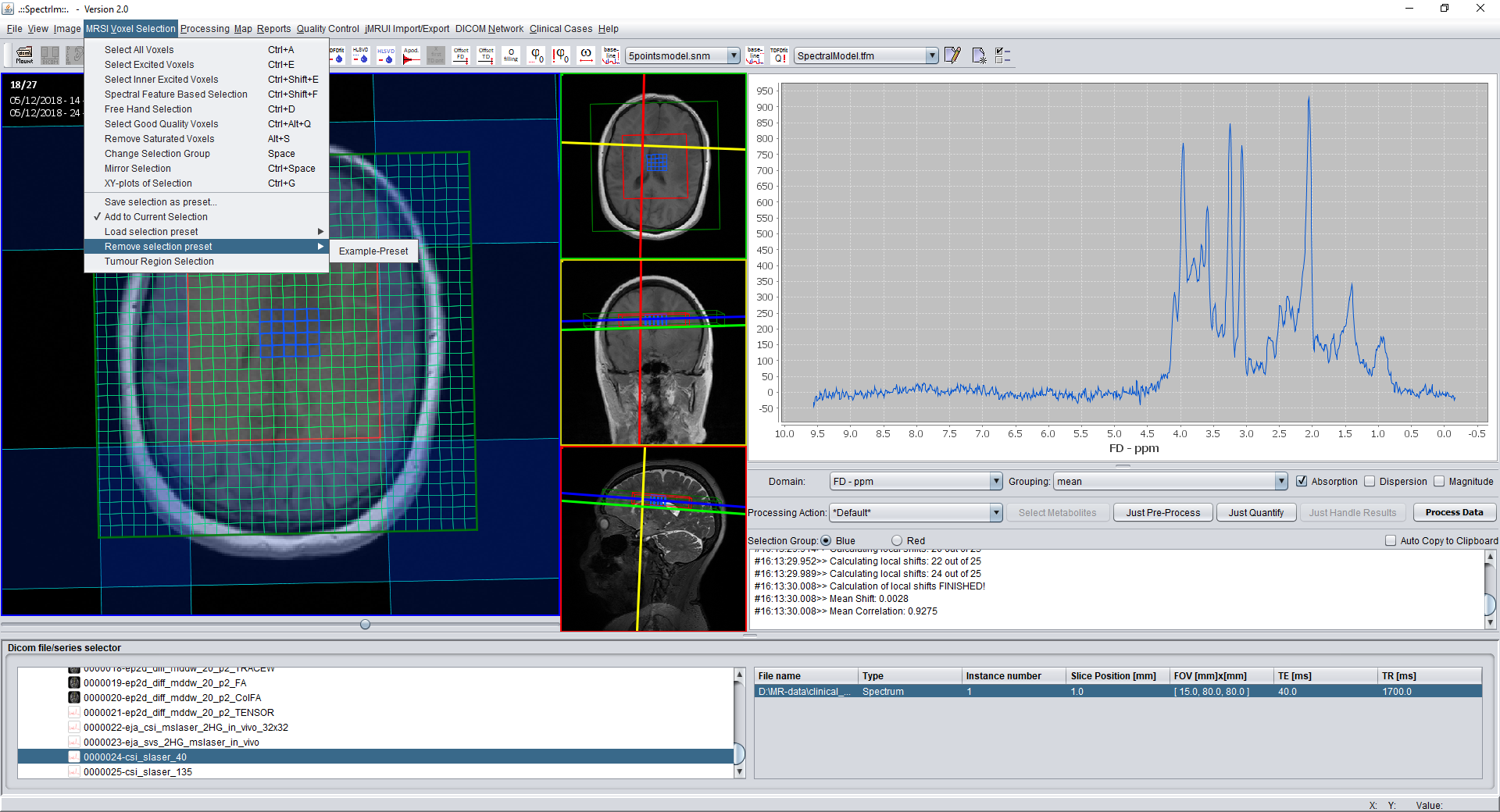
The menu item disappears only as a result, and the user should manually update to make the preset disappear from the spectral grid as well (as was also in the example above).
spectrIm-QMRS -> MRSI Voxel Selection menu-> Tumor region selection
As a result of selecting this menu item, a popup frame will appear allowing the user to define manually selected regions and assign them to different tissue classes. This is handy functionalty for definition of ground truths in tumor diagnostics and machine learning.
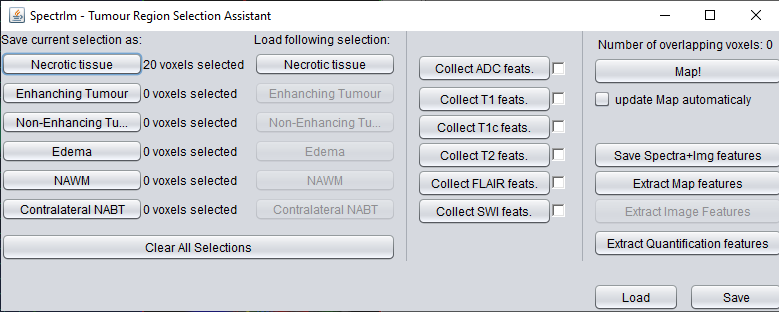
How to use this GUI will be topic of a video which --> Under construction <--