The spectrIm-QMRS file menu
The file menu enables manual file loading/saving import output as well as input and output of different types of files.
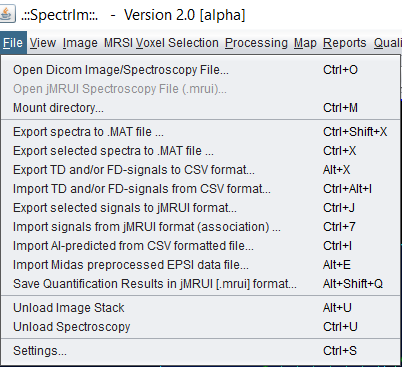
The following menu items are available:
spectrIm-QMRS ->File menu -> Open DICOM Image/Spectroscopy data ..
This menu enables the selection of a single dicom file from the file system. For the DICOM standard please refer to DICOM Image/Spectroscopy data formats.
spectrIm-QMRS -> File menu -> Open jMRUI Spectroscopy data ..
This menu enables the of a single jMRUI spectroscopy file (.mrui) from the file system.
This function is in the current version disabled.
spectrIm-QMRS -> File menu -> Mount directory ..
This menu enables the user to mount a filesystem directory in which DICOM data are stored. spectrIm-QMRS will parse the directory and all subdirectories, and builds up a tree structures on study level, patient level, series level makes the DICOM files directy asssisble (readable) to human users.
spectrIm-QMRS -> File menu -> Export data to .MAT file ..
This menu enables the export of the selected spectra to a .MAT file system allowing for further analysis of the data in Matlab.
spectrIm-QMRS -> File menu -> Export data to .MAT file ..
This menu enables the export of all spectra to a .MAT file system allowing for further analysis of the data in Matlab.
spectrIm-QMRS -> File menu -> Export data to .CSV file ..
This menu enables the export of all spectra to a .CSV file allowing for further (statistic) analysis of the data in or others.
spectrIm-QMRS -> File menu -> Import data from .CSV file ..
This menu enables the import of spectra from a .CSV formatted file allowing for further analysis/display of the data in spectrIm-QMRS. The CVS-typed data can however only be imported if a data file was already imported before. The FD spectral data is organized such that each row contains one complex valued spectrum. The first element indicates the 3D locater string which has the following format: "x_y_z" in which x,y,z are integers, which count from the top left point which is "0_0_0". This string denoted as "voxel position association string". Note that the x-direction is from left to right, the y-direction from the top down, and the z-direction are the number of MRSI slices of a 3D dataset. In 2D MRSI dataset only x and y are set as defined above, and z=0. The string "x_y_z" is used throughout spectrIm-QMRS as an association string which can link (extermal) data to a specific spectrum in a multidimensional dataset.
Example: 12_17_0 indicates the 13th voxel from the left and the 18th voxel from the top, in slice 0.
spectrIm-QMRS -> File menu -> Export selected spectra to jMRUI Spectroscopy formatted (.mrui) data file ..
This menu enables the export of selected spectra to a single jMRUI spectroscopy file (.mrui) in the local file system.
spectrIm-QMRS -> File menu -> Import signals from jMRUI format (association) ..
This menu enables the import of jMRUI formatted spectroscopy files (.mrui) from the file system. The signal indicators in the jmrui files must however contain a so-called "voxel position association string". This string denotes the 3D locater string which should have the following format: "x_y_z" in which x,y,z are integers, which count from the top left point which is "0_0_0". Note that the x-direction is from left to right, the y-direction from the top down, and the z-direction are the number of MRSI slices of a 3D dataset. In 2D MRSI dataset only x and y are set as defined above, and z=0. The string "x_y_z" is used throughout spectrIm-QMRS as an association string which can link (extermal) data to a specific spectrum in a multidimensional dataset.
Example: 12_17_0 indicates the 13th voxel from the left and the 18th voxel from the top, in slice 0.
spectrIm-QMRS -> File menu -> Import AI predicted [parameters] from CSV formatted file ..
This menu enables the import AI predicted [parameters] from CSV formatted file in the file system. This is an option if an external machine learning, deep learning or NLLS quantification has been used. The .CSV should be organized such that the quantification results of one spectrum should be organized row-wise and the first element of each row should be the "voxel position association string". This means that a dataset must already be loaded and after this, an associated result file can be loaded which is associated to the already loaded datafile.
spectrIm-QMRS -> File menu -> Import Midas preprocessed EPSI data from .sid file ..
This menu enables the import of an .sid formatted data file generated with the MIDAS software. This case is a special case, since a DICOM formatted spectroscopic image stack must be loaded first before the .sid file can be loaded. From the 3D spectroscopic image the geometrical DICOM-tags (slice location, slice orientation) are loaded and enables the proper display of the EPSI-MRSI data in the coordinate system of the examination. After the load of this two datasets (first the spectroscopic image, and the .sid file) any other high resolution MRI-image stack can be displayed for visually inspect spectra at well defined anatomic locations.
spectrIm-QMRS -> File menu -> Save quantification results in jMRUI [.mrui] format ..
This menu enables the export of quantification results obtained with spectrIm-QMRS into jMRUI format. The results can than further be analyzed in jMRUI. The "voxel position association string" is added to each individual voxel of the MRSI dataset. This string denotes the 3D locater string which should have the following format: "x_y_z" in which x,y,z are integers, which count from the top left point which is "0_0_0". Note that the x-direction is from left to right, the y-direction from the top down, and the z-direction are the number of MRSI slices of a 3D dataset. In 2D MRSI dataset only x and y are set as defined above, and z=0. The string "x_y_z" is used throughout spectrIm-QMRS as an association string which can link (extermal) data to a specific spectrum in a multidimensional dataset.
Example: 12_17_0 indicates the 13th voxel from the left and the 18th voxel from the top, in slice 0.
spectrIm-QMRS -> File menu -> Unload image stack [data]
Unloads the background image stack in the image viewer with spectral grid overlays. If an MRS(I) dataset was loaded, this data remain in memory.
spectrIm-QMRS -> File menu -> Unload spectroscopy [data]
Unloads the background spectroscopy data from spectrIm-QMRS. If an image data stack was loaded, this data remain in memory and visible in the viewer.
spectrIm-QMRS -> File menu -> Settings [of spectrIm-QMRS]
Launches a GUI which enables the definition of e.g. processing pipelines that can be used within spectrIm-QMRS. There are however also different GUIs which enable the user other settings to be used in spectrIm-QMRS.